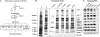Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth - PubMed (original) (raw)
Comparative Study
. 2007 Mar;27(5):1889-903.
doi: 10.1128/MCB.01506-06. Epub 2006 Dec 18.
Affiliations
- PMID: 17178841
- PMCID: PMC1820476
- DOI: 10.1128/MCB.01506-06
Comparative Study
Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth
Irina Issaeva et al. Mol Cell Biol. 2007 Mar.
Abstract
ALR (MLL2) is a member of the human MLL family, which belongs to a larger SET1 family of histone methyltransferases. We found that ALR is present within a stable multiprotein complex containing a cohort of proteins shared with other SET1 family complexes and several unique components, such as PTIP and the jumonji family member UTX. Like other complexes formed by SET1 family members, the ALR complex exhibited strong H3K4 methyltransferase activity, conferred by the ALR SET domain. By generating ALR knockdown cell lines and comparing their expression profiles to that of control cells, we identified a set of genes whose expression is activated by ALR. Some of these genes were identified by chromatin immunoprecipitation as direct ALR targets. The ALR complex was found to associate in an ALR-dependent fashion with promoters and transcription initiation sites of target genes and to induce H3K4 trimethylation. The most characteristic features of the ALR knockdown cells were changes in the dynamics and mode of cell spreading/polarization, reduced migration capacity, impaired anchorage-dependent and -independent growth, and decreased tumorigenicity in mice. Taken together, our results suggest that ALR is a transcriptional activator that induces the transcription of target genes by covalent histone modification. ALR appears to be involved in the regulation of adhesion-related cytoskeletal events, which might affect cell growth and survival.
Figures
FIG. 1.
Purification of the ALR complex and identification of ALR-associated proteins. (A) Scheme for four-step purification of the ALR complex. IAC, immunoaffinity column. (B) Coomassie staining of proteins eluted from anti-ALR, anti-PTIP, anti-UTX, and control immunoaffinity columns. About 50 μg of affinity-purified complex was eluted from each IAC (except for the control column); 98% of this amount was used for Coomassie staining and subsequent mass spectrometry analysis. For comparison of protein pattern complexities before, during, and after complex purification, Coomassie staining patterns of 50 μg of crude K562 nuclear extract (lane 1) and 50 μg of the ALR-enriched Superose 6 pooled fractions (lane 2) are shown, corresponding to 1/6,000 and 1/60, respectively, of the total material used for complex purification and subjected to IAC. The positions of molecular mass markers are indicated on the left. *, components of SET1-like histone methyltransferase complexes. (C) The identities of ALR complex components were verified by immunoblotting.
FIG. 2.
Confirmation of ALR complex composition. (A) Size fractionation of the enriched ALR complex. The ALR-enriched K562 nuclear extracts, obtained from subsequent purification steps on P11 and Q-Sepharose columns, were applied to a preparative Superose 6 column. One percent of each fraction collected from Superose 6 was analyzed by immunoblotting using Abs directed to various ALR complex components. fr#, fraction number. The positions of molecular mass markers determined in a parallel run are indicated at the top. (B) Coprecipitation of ALR with other ALR complex components from A549 cells. Crude nuclear extracts obtained from A549 cells were utilized in reciprocal coimmunoprecipitations. Proteins coprecipitated (IP) with anti-ALR, anti-PTIP, anti-UTX, and anti-RBQ3 Abs, as well as with control IgG, were separated on SDS-PAGE and immunoblotted (IB) with the indicated Abs. Since different amounts of Abs were used in some IPs, the analysis is qualitative rather than quantitative.
FIG. 3.
Histone methyltransferase activities of the ALR complex and of the ALR SET domain. (A) The ALR complex methylates histone H3. Anti-ALR, anti-PTIP, anti-UTX, or control immunoprecipitate (IP) was incubated with core histones and the methyl donor H3-SAM. Samples were resolved on 15% SDS-PAGE, stained with Coomassie blue (right), amplified, dried, and fluorographed (left). Histone subunits are indicated on the right. (B) ALR SET domain methylates H3K4. Two ALR SET domain-containing fragments, encompassing the ALR amino acids 5009 to 5261 and 5089 to 5261 (designated SET a and SET b, respectively), were fused to GST and overexpressed in 293T cells. Cells overexpressing GST only were used as a control. The overexpressed polypeptides were immunoprecipitated from cell lysates using anti-GST Ab and incubated with N-terminal H3 peptides containing unmodified K4 and K9, dimethylated K4, dimethylated K9, or trimethylated K9. Samples were resolved on 20% SDS-PAGE, stained with Coomassie blue (bottom), and fluorographed (top). (C) The recombinant ALR SET domain does not interact with the ASH2, RBQ3, and WDR5 proteins. GST-SET fusions or control GST was precipitated from 293T cell lysates with anti-GST Ab and analyzed by immunoblotting using anti-GST, -ASH2, -RBQ3, or -WDR5 Abs. Five percent of each GST-SET- or control GST-overexpressing lysate was loaded as an input control. (D) The ALR complex methylates H3 peptides containing either unmodified or dimethylated K4. Anti-ALR, anti-PTIP, anti-UTX, or control immunoprecipitate was incubated with unmodified or appropriately methylated H3 peptides. The reaction was analyzed as described above.
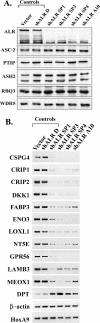
FIG. 4.
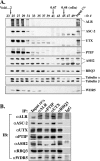
(A) Effects of ALR shRNAs on the level of ALR. Four stable cell lines generated by infection of HeLa cells with retroviruses carrying four different ALR shRNA constructs (lanes shALR SP1 to shALR A10) and two control HeLa cell lines, infected with the retroviral vector or with noncompetent ALR shRNA D, were examined by immunoblotting for expression of the ALR protein and some ALR complex components. (B) Confirmation of the microarray results by RT-PCR analysis. RNAs isolated from the above-described cell lines were used for semiquantitative RT-PCR analysis to monitor the expression of selected genes. PCR cycle numbers were individually optimized for each gene so that each reaction fell into the linear range of product amplification. The genes analyzed included 11 downregulated genes (CSPG4, CRIP1, CRIP2, DKK1, FABP3, ENO3, LOXL1, NT5E, GPR56, LAMB3, and MEOX1), 1 upregulated gene (DPT), and 2 control genes (β-actin and HoxA9).
FIG. 5.
ChIP analysis of CRIP2 (A), ENO3 (B), CSPG4 (C), DKK1 (D), and HoxA9 (E) loci in vector-infected and SP3 cells. The presence of the ALR, PTIP, and UTX proteins, as well as of histone H3K4 trimethylation, on several regions of CRIP2, ENO3, CSPG4, and DKK1 loci and on the 5′ end of the HoxA9 locus was examined. Chromatin was prepared from the control vector-infected cells and from the ALR-deprived SP3 cell line. The analyzed portions of the genes are depicted to the right of panels A to D and at the bottom of panel E. The boxes shown on the schemes indicate the first exons, and the dark areas correspond to the coding regions. The arrows point to the transcription initiation sites; a to e correspond to the sequences amplified by PCR following immunoprecipitation with each of the six Abs (two anti-ALR Abs, anti-PTIP, anti-UTX, control IgG, and anti-trimethylated H3K4). For input, 0.1% or 0.5% of the amount of sonicated chromatin to be subsequently processed for ChIP was removed and PCR amplified.
FIG. 6.
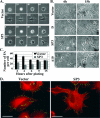
Changes in the morphology of ALR knockdown cells during spreading. (A and B) Differences in dynamics and modes of cell spreading. (A) Control (Vector) and ALR-deficient (SP3) cells were plated on fibronectin-coated dishes and analyzed by time-lapse photography during the subsequent 4 h. The numbers represent the time after plating in min. (B) Two control (Vector and D) and two ALR-underexpressing (SP3 and A10) cell lines were seeded on fibronectin-coated coverslips and fixed with 3.7% paraformaldehyde at different time points during the subsequent 15 h. Typical photographs of the cells at 6 and 15 h after being plated are shown. (C and D) Decreased numbers of FA and alterations in actin cytoskeleton organization in spreading ALR-deficient cells. (C) Control (Vector) and ALR knockdown (SP3) cells were processed as described for panel B, and FA were double stained with anti-vinculin and anti-paxillin Abs. The numbers of FA formed by each cell type at the indicated time points were calculated. The data represent the average FA number of 30 cells plus standard deviation. (D) The cells described above, fixed at 3 h after being plated, were stained for filamentous actin with tetramethyl rhodamine isothiocyanate-phalloidin (red) and for FA with anti-paxillin Ab (green). Bars (A, B, and D), 50 μm.
FIG. 7.
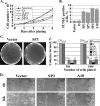
ALR knockdown affects cell growth, apoptosis, and migration. (A) Cell growth assay. The growth rates of three ALR-deprived HeLa cell lines (SP3, SP4, and A10) and of the control cells (Vector and D) were determined by plating 105 cells in triplicate on six-well plates and counting the cells every 24 h during the subsequent 4 days. The error bars represent the standard deviations of three independent experiments. (B) Increased apoptosis in the ALR-deprived cells. Control (Vector and D) and ALR-deprived (SP1, SP3, SP4, and A10) HeLa cells were stained with the TUNEL assay kit. Percentages of TUNEL-positive (apoptotic) populations are shown. The error bars represent the standard deviations of three independent experiments. (C) ALR knockdown inhibits soft-agar growth of HeLa cells. Five hundred and 1,000 cells/3-cm dish were seeded on 0.3% agar in triplicate. Three weeks later, the dishes were photographed and the colonies were counted. The experiment was repeated twice. Typical photographs of plates seeded with control (Vector) and ALR-deficient (SP3) cells are shown on the left. The histogram shows the numbers of colonies formed in each cell line at both plating concentrations as a percentage of the control vector-expressing cells. Absolute colony numbers are shown above the bars. (D) Effect of ALR knockdown on cell migration. A scratch injury was introduced with a micropipette tip into confluent cultures of the control (Vector) and ALR-depleted (SP3) A549 cells. Phase-contrast images of cultures were taken either immediately after scratching (0 h) or 16 h later.
FIG. 8.
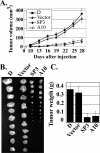
Decreased tumorigenicity of ALR knockdown cells in nude mice. (A) Ten animals in each group were injected subcutaneously with 106 cells of either control (Vector and D) or ALR-deficient (SP3 and A10) HeLa cell lines. Tumor volumes were determined at regular time intervals. The experiment was stopped after 4 weeks, when the tumors in the control groups reached 1 to 1.5 cm in diameter. The results represent the means ± standard errors (SE). (B) Photographs and (C) mean weights of tumors formed in each group at 4 weeks after injection. Error bars, SE.
Similar articles
- The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex.
Patel SR, Kim D, Levitan I, Dressler GR. Patel SR, et al. Dev Cell. 2007 Oct;13(4):580-92. doi: 10.1016/j.devcel.2007.09.004. Dev Cell. 2007. PMID: 17925232 Free PMC article. - Fgfr3 is a transcriptional target of Ap2delta and Ash2l-containing histone methyltransferase complexes.
Tan CC, Walsh MJ, Gelb BD. Tan CC, et al. PLoS One. 2009 Dec 31;4(12):e8535. doi: 10.1371/journal.pone.0008535. PLoS One. 2009. PMID: 20046871 Free PMC article. - Pax7 activates myogenic genes by recruitment of a histone methyltransferase complex.
McKinnell IW, Ishibashi J, Le Grand F, Punch VG, Addicks GC, Greenblatt JF, Dilworth FJ, Rudnicki MA. McKinnell IW, et al. Nat Cell Biol. 2008 Jan;10(1):77-84. doi: 10.1038/ncb1671. Epub 2007 Dec 9. Nat Cell Biol. 2008. PMID: 18066051 Free PMC article. - Structure, Activity and Function of the MLL2 (KMT2B) Protein Lysine Methyltransferase.
Klonou A, Chlamydas S, Piperi C. Klonou A, et al. Life (Basel). 2021 Aug 12;11(8):823. doi: 10.3390/life11080823. Life (Basel). 2021. PMID: 34440566 Free PMC article. Review. - Cancer-epigenetic function of the histone methyltransferase KMT2D and therapeutic opportunities for the treatment of KMT2D-deficient tumors.
Dhar SS, Lee MG. Dhar SS, et al. Oncotarget. 2021 Jun 22;12(13):1296-1308. doi: 10.18632/oncotarget.27988. eCollection 2021 Jun 22. Oncotarget. 2021. PMID: 34194626 Free PMC article. Review.
Cited by
- Coexpression of nuclear receptors and histone methylation modifying genes in the testis: implications for endocrine disruptor modes of action.
Anderson AM, Carter KW, Anderson D, Wise MJ. Anderson AM, et al. PLoS One. 2012;7(4):e34158. doi: 10.1371/journal.pone.0034158. Epub 2012 Apr 4. PLoS One. 2012. PMID: 22496781 Free PMC article. - Report of the First Clinical Case of a Moroccan Kabuki Patient with a Novel MLL2 Mutation.
Ratbi I, Fejjal N, Micale L, Augello B, Fusco C, Lyahyai J, Merla G, Sefiani A. Ratbi I, et al. Mol Syndromol. 2013 Mar;4(3):152-6. doi: 10.1159/000346798. Epub 2013 Jan 30. Mol Syndromol. 2013. PMID: 23653588 Free PMC article. - Epigenomic Regulation of Schwann Cell Reprogramming in Peripheral Nerve Injury.
Ma KH, Hung HA, Svaren J. Ma KH, et al. J Neurosci. 2016 Aug 31;36(35):9135-47. doi: 10.1523/JNEUROSCI.1370-16.2016. J Neurosci. 2016. PMID: 27581455 Free PMC article. - Requirement for MLL3 in p53 regulation of hepatic expression of small heterodimer partner and bile acid homeostasis.
Kim DH, Kim J, Lee JW. Kim DH, et al. Mol Endocrinol. 2011 Dec;25(12):2076-83. doi: 10.1210/me.2011-1198. Epub 2011 Oct 27. Mol Endocrinol. 2011. PMID: 22034226 Free PMC article. - Driver mutations of cancer epigenomes.
Roy DM, Walsh LA, Chan TA. Roy DM, et al. Protein Cell. 2014 Apr;5(4):265-96. doi: 10.1007/s13238-014-0031-6. Epub 2014 Mar 14. Protein Cell. 2014. PMID: 24622842 Free PMC article.
References
- Campoli, M. R., C. C. Chang, T. Kageshita, X. Wang, J. B. McCarthy, and S. Ferrone. 2004. Human high molecular weight-melanoma-associated antigen (HMW-MAA): a melanoma cell surface chondroitin sulfate proteoglycan (MSCP) with biological and clinical significance. Crit. Rev. Immunol. 24:267-296. - PubMed
- Chen, J. J., K. Peck, T. M. Hong, S. C. Yang, Y. P. Sher, J. Y. Shih, R. Wu, J. L. Cheng, S. R. Roffler, C. W. Wu, and P. C. Yang. 2001. Global analysis of gene expression in invasion by a lung cancer model. Cancer Res. 61:5223-5230. - PubMed
- Chu, Y. W., P. C. Yang, S. C. Yang, Y. C. Shyu, M. Hendrix, R. Wu, and C. W. Wu. 1997. Selection of invasive and metastatic subpopulations from a human lung adenocarcinoma cell line. Am. J. Respir. Cell Mol. Biol. 17:353-360. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous