p14-MP1-MEK1 signaling regulates endosomal traffic and cellular proliferation during tissue homeostasis - PubMed (original) (raw)
. 2006 Dec 18;175(6):861-8.
doi: 10.1083/jcb.200607025.
Nicole Taub, Robert Kurzbauer, Diana Hilber, Mariana E de Araujo, Miriam Erlacher, Martin Offterdinger, Andreas Villunger, Stephan Geley, Georg Bohn, Christoph Klein, Michael W Hess, Lukas A Huber
Affiliations
- PMID: 17178906
- PMCID: PMC2064696
- DOI: 10.1083/jcb.200607025
p14-MP1-MEK1 signaling regulates endosomal traffic and cellular proliferation during tissue homeostasis
David Teis et al. J Cell Biol. 2006.
Abstract
The extracellular signal-regulated kinase (ERK) cascade regulates proliferation, differentiation, and survival in multicellular organisms. Scaffold proteins regulate intracellular signaling by providing critical spatial and temporal specificity. The scaffold protein MEK1 (mitogen-activated protein kinase and ERK kinase 1) partner (MP1) is localized to late endosomes by the adaptor protein p14. Using conditional gene disruption of p14 in mice, we now demonstrate that the p14-MP1-MEK1 signaling complex regulates late endosomal traffic and cellular proliferation. This function its essential for early embryogenesis and during tissue homeostasis, as revealed by epidermis-specific deletion of p14. These findings show that endosomal p14-MP1-MEK1 signaling has a specific and essential function in vivo and, therefore, indicate that regulation of late endosomal traffic by extracellular signals is required to maintain tissue homeostasis.
Figures
Figure 1.
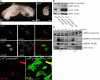
p14 is an essential gene required for embryonic development and endosomal ERK activation. (A) Homozygous p14 −/− embryos die around gastrulation. Representative p14 −/− and p14 −/+ E8 embryos are shown. (B) p14 f/− and p14 −/− MEFs were infected with either control retrovirus (IRES-GFP) or p14 retrovirus (p14-IRES-GFP). p14 f/−;IRES-GFP, p14 −/−;IRES-GFP, p14 −/−;p14-IRES-GFP, and p14 f/−;p14-IRES-GFP cell lysates were separated by SDS-PAGE, analyzed by Western blotting, and probed with the indicated antibodies. The asterisk marks the background band of the p14 antibody. (C) p14 f/− and p14 −/− MEFs were infected with myc-MP1 retrovirus. myc-MP1 (green) colocalizes with LAMP1 (red) in p14 f/− (top) and mislocalizes to the cytoplasm in p14 −/− MEFs (middle). p14 −/− MEFs were infected with p14-IRES-GFP retrovirus (green), transfected with myc-MP1 (red), and analyzed by immunofluorescence microscopy using anti-myc antibodies. (bottom) (D) p14 f/− and p14 −/− MEFs were starved overnight and stimulated with 100 ng/ml EGF for the indicated times. Cell lysates were separated by SDS-PAGE, analyzed by Western blotting, and probed with the indicated antibodies. Bars (A), 300 μm; (C), 10 μm.
Figure 2.
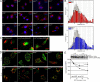
p14 is required for late endosomal positioning and EGFR transport. (A) p14 f/− and p14 −/− as well as MEK1 −/− and KSR1 −/− MEFs were subjected to immunofluorescence analysis with the indicated antibodies. EEA1, LAMP1, and LBPA are shown in red, and nuclei are stained with DAPI (blue). (B) Distribution analysis of LAMP1-positive endosomes in p14 f/− and p14 −/− MEK1 +/+ and MEK1 −/− MEFs. The distance distribution of LAMP1-positive endosomes to the nucleus is shown. (C) p14 f/− were infected with control retrovirus (IRES-GFP), and p14 −/− MEFs were infected with retrovirus-expressing p14 (p14;IRES-GFP) or p14caax (p14caax;IRES-GFP) and subjected to immunofluorescence analysis. LAMP1 is shown in red, and GFP expression from different IRES-GFP retroviruses indicates p14 or p14caax expression. (D) p14 f/−, p14 −/−, MEK1 −/−, and KSR1 −/− MEFs were starved overnight and stimulated with 100 ng/ml of fluorescently labeled EGF. Cells were fixed at the indicated times and subjected to confocal immunofluorescence analysis with the indicated antibodies. EGF is shown in green, and EEA1 and LAMP1 are shown in red. Colocalization of EGF with either EEA1 or LAMP1 is shown in yellow (arrows). (E) p14 f/− and p14 −/− MEFs were starved overnight and stimulated with 100 ng/ml EGF for the indicated times. Cell lysates were separated by SDS-PAGE, analyzed by Western blotting, and probed with the indicated antibodies. One representative EGFR immunoblot is shown. The EGFR degradation was analyzed in three independent experiments, and EGFR protein levels were normalized to total ERK protein levels. The graph shows the mean EGFR degradation in p14 f/− and p14 −/− MEFs. Error bars represent SD. Bars, 10 μm.
Figure 3.
The p14–MP1 complex is required for epidermal development and regulates ERK signaling and EGFR degradation. (A) Representative phenotype of E18.5 p14Δep/+ and p14Δep embryos. Semithin sections of E18.5 p14Δep and p14Δep/+ skin were stained with Toluidine blue. The p14Δep/+ skin consists of stratum corneum (SC), granular layers (GL), spinous layers (SL), and the basal layer (BL). p14Δep lacks the stratum corneum and granular layer. The arrow indicates a nucleated cell in the uppermost layer of p14Δep skin. (B) E18.5 p14Δep and p14Δep/+ embryos were subjected to an epidermal barrier assay. X-Gal fully penetrated the p14Δep epidermis of embryos. (C–E) Frozen skin sections from E18.5 p14Δep and p14Δep/+ embryos were analyzed by confocal laser-scanning immunofluorescence microscopy. β4-Integrin is shown in red. Bars, 10 μm. (C) pERK1/2 (green) is strongly reduced in the basal compartment of p14Δep epidermis. Note the unspecific background staining in the stratum corneum in panel p14Δep/+. (D) EGFR (green) is present at the plasma membrane of suprabasal keratinocytes in the p14Δep epidermis. Insets show magnifications of boxed areas. (E) Keratin 6 is shown in green and is normally confined to the innermost cell layer of the outer root sheath. (F) Equal protein amounts of epidermal lysates from E18.5 p14Δep and p14Δep/+ embryos were analyzed by Western blotting with the indicated antibodies. Asterisks indicate the background bands of the p14 and MP1 antibodies.
Figure 4.
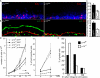
The p14–MP1 complex regulates cell cycle progression and cellular proliferation. (A and B) Frozen skin sections from E18.5 p14Δep and p14Δep/+ embryos were analyzed by confocal immunofluorescence microscopy and quantification. Representative images are shown. Bars, 10 μm. (A) DAPI is shown in blue, and BrdU is shown in red. The dotted white line indicates the epidermal–dermal boundary. (B) β4-Integrin is shown in red, and phosphohistone H3 is shown in green. Note the unspecific background staining in the stratum corneum in panel p14Δep/+. (C) BrdU-positive cells were quantified in 25 random fields of view from four different embryos. In the p14Δep/+ epidermis, 22 ± 2.2 (SD [error bars]) cells/field of view were BrdU-positive cells. In the p14Δep epidermis, 12 ± 1.51 (54.5%) were BrdU positive. Phosphohistone H3 (pH3)–positive cells were quantified in 58 random fields of view from six different embryos. The mean per field of view was 10 ± 1.1 phosphohistone H3–positive cells (100%) in the p14Δep/+ epidermis and 5 ± 0.57 (50%) in the p14Δep epidermis. P < 0.001. (D) 0.5 × 104 p14 f/−;IRES-GFP, p14 −/−;IRES-GFP, p14 −/−;p14-IRES-GFP, and p14 −/−;p14caax-IRES-GFP MEFs were plated. Cells were counted at the indicated times. On day 4, p14 f/−; IRES-GFP had grown to 2.02 × 104 MEFs (100 ± 21%), p14 −/−;IRES-GFP had grown to 1.104 MEFs (54.4 ± 8%), p14 −/−;p14-IRES-GFPhad grown to 2.104 MEFs (103 ± 16%), and p14 −/−;p14caax-IRES-GFP had grown to 0.9 × 104 MEFs (44.5 ± 22%; n = 3). Growth-arrested MEFs were released into S phase. The number of mitotic cells was analyzed by DAPI and phosphohistone H3 staining. Mitotic indexes before the release (t = 0) were as follows: p14 f/−;IRES-GFP, 5.2 ± 0.2% (n = 166); p14 −/−;IRES-GFP, 3.8 ± 1.8% (n = 300); p14 −/−;p14-IRES-GFP, 5.2 ± 0.8% (n = 325); and p14 −/−;p14caax-IRES-GFP, 1.4 ± 0.2% (n = 260). 24 h after the release (t = 24 h), mitotic indexes were as follows: p14 f/−;IRES-GFP, 24 ± 4.5% (n = 122); p14 −/−;IRES-GFP, 6 ± 1.7% (n = 83); p14 −/−;p14-IRES-GFP, 21 ± 6.6% (n = 109); and p14 −/−;p14caax-IRES-GFP, 3% (n = 42). (E) Quantitative analysis by propidium iodide FACS of the DNA content of p14 f/− and p14 −/− MEFs 6 h after mitogenic stimulation. Results for p14 f/− and p14 −/− MEFs, respectively, were as follows: G1 phase, 42.2 and 70%; S phase, 44.2 and 17.7%; and G2–M phase, 12.8 and 12.2% (n = 3).
Similar articles
- Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction.
Teis D, Wunderlich W, Huber LA. Teis D, et al. Dev Cell. 2002 Dec;3(6):803-14. doi: 10.1016/s1534-5807(02)00364-7. Dev Cell. 2002. PMID: 12479806 - Crystal structure of the p14/MP1 scaffolding complex: how a twin couple attaches mitogen-activated protein kinase signaling to late endosomes.
Kurzbauer R, Teis D, de Araujo ME, Maurer-Stroh S, Eisenhaber F, Bourenkov GP, Bartunik HD, Hekman M, Rapp UR, Huber LA, Clausen T. Kurzbauer R, et al. Proc Natl Acad Sci U S A. 2004 Jul 27;101(30):10984-9. doi: 10.1073/pnas.0403435101. Epub 2004 Jul 19. Proc Natl Acad Sci U S A. 2004. PMID: 15263099 Free PMC article. - Interactions between kinase scaffold MP1/p14 and its endosomal anchoring protein p18.
Magee J, Cygler M. Magee J, et al. Biochemistry. 2011 May 10;50(18):3696-705. doi: 10.1021/bi101972y. Epub 2011 Apr 15. Biochemistry. 2011. PMID: 21452851 - The MEK1 scaffolding protein MP1 regulates cell spreading by integrating PAK1 and Rho signals.
Pullikuth A, McKinnon E, Schaeffer HJ, Catling AD. Pullikuth A, et al. Mol Cell Biol. 2005 Jun;25(12):5119-33. doi: 10.1128/MCB.25.12.5119-5133.2005. Mol Cell Biol. 2005. PMID: 15923628 Free PMC article. - Metabolic regulation through the endosomal system.
Gilleron J, Gerdes JM, Zeigerer A. Gilleron J, et al. Traffic. 2019 Aug;20(8):552-570. doi: 10.1111/tra.12670. Epub 2019 Jun 24. Traffic. 2019. PMID: 31177593 Free PMC article. Review.
Cited by
- p38 signaling and receptor recycling events in a microfluidic endothelial cell adhesion assay.
Vickers DA, Chory EJ, Harless MC, Murthy SK. Vickers DA, et al. PLoS One. 2013 Jun 7;8(6):e65828. doi: 10.1371/journal.pone.0065828. Print 2013. PLoS One. 2013. PMID: 23762436 Free PMC article. - LAMTOR1 depletion induces p53-dependent apoptosis via aberrant lysosomal activation.
Malek M, Guillaumot P, Huber AL, Lebeau J, Pétrilli V, Kfoury A, Mikaelian I, Renno T, Manié SN. Malek M, et al. Cell Death Dis. 2012 Apr 19;3(4):e300. doi: 10.1038/cddis.2012.39. Cell Death Dis. 2012. PMID: 22513874 Free PMC article. - The late endosomal adaptor molecule p14 (LAMTOR2) represents a novel regulator of Langerhans cell homeostasis.
Sparber F, Scheffler JM, Amberg N, Tripp CH, Heib V, Hermann M, Zahner SP, Clausen BE, Reizis B, Huber LA, Stoitzner P, Romani N. Sparber F, et al. Blood. 2014 Jan 9;123(2):217-27. doi: 10.1182/blood-2013-08-518555. Epub 2013 Oct 3. Blood. 2014. PMID: 24092934 Free PMC article. - Biogenesis of lysosome-related organelles complex-1 (BORC) regulates late endosomal/lysosomal size through PIKfyve-dependent phosphatidylinositol-3,5-bisphosphate.
Yordanov TE, Hipolito VEB, Liebscher G, Vogel GF, Stasyk T, Herrmann C, Geley S, Teis D, Botelho RJ, Hess MW, Huber LA. Yordanov TE, et al. Traffic. 2019 Sep;20(9):674-696. doi: 10.1111/tra.12679. Traffic. 2019. PMID: 31314175 Free PMC article.
References
- Acharya, U., A. Mallabiabarrena, J.K. Acharya, and V. Malhotra. 1998. Signaling via mitogen-activated protein kinase kinase (MEK1) is required for Golgi fragmentation during mitosis. Cell. 92:183–192. - PubMed
- Galabova-Kovacs, G., A. Kolbus, D. Matzen, K. Meissl, D. Piazzolla, C. Rubiolo, K. Steinitz, and M. Baccarini. 2006. ERK and beyond: insights from B-Raf and Raf-1 conditional knockouts. Cell Cycle. 5:1514–1518. - PubMed
- Giroux, S., M. Tremblay, D. Bernard, J.F. Cardin-Girard, S. Aubry, L. Larouche, S. Rousseau, J. Huot, J. Landry, L. Jeannotte, and J. Charron. 1999. Embryonic death of Mek1-deficient mice reveals a role for this kinase in angiogenesis in the labyrinthine region of the placenta. Curr. Biol. 9:369–372. - PubMed
- Hardman, M.J., P. Sisi, D.N. Banbury, and C. Byrne. 1998. Patterned acquisition of skin barrier function during development. Development. 125:1541–1552. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous