Characterization of multiple multivesicular body sorting determinants within Sna3: a role for the ubiquitin ligase Rsp5 - PubMed (original) (raw)
Characterization of multiple multivesicular body sorting determinants within Sna3: a role for the ubiquitin ligase Rsp5
Andrea J Oestreich et al. Mol Biol Cell. 2007 Feb.
Abstract
A subset of proteins that transit the endosomal system are directed into the intralumenal vesicles of multivesicular bodies (MVBs). MVB formation is critical for a variety of cellular functions including receptor down-regulation, viral budding, antigen presentation, and the generation of lysosome-related organelles. Entry of transmembrane proteins into the intralumenal vesicles of a MVB is a highly regulated process that is positively modulated by covalent modification of cargoes with ubiquitin. To identify additional MVB sorting signals, we examined the previously described ubiquitination-independent MVB cargo Sna3. Although Sna3 ubiquitination is not essential, Sna3 MVB sorting is positively modulated by its ubiquitination. Examination of MVB sorting determinants within a form of Sna3 lacking all lysine residues identified two critical regions: an amino-terminal tyrosine-containing region and a carboxyl-terminal PPAY motif. This PPAY motif interacts with the WW domains of the ubiquitin ligase Rsp5, and mutations in either the WW or, surprisingly, the HECT domains of Rsp5 negatively impacted MVB targeting of lysine-minus Sna3. These data indicate that Rsp5 function is required for MVB targeting of Sna3 in a capacity beyond cargo ubiquitination. These results uncover a series of determinants impacting Sna3 MVB sorting, including unexpected roles for Rsp5.
Figures
Figure 1.
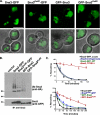
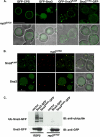
Sna3 ubiquitination is not a requisite for MVB targeting. (A) GFP-tagged forms of Sna3 were expressed in _sna3_Δ cells and visualized by fluorescence microscopy. Sna3KallR denotes a mutant form of Sna3 in which all lysine residues have been mutated to arginines. The position of the GFP tag relative to Sna3 is indicated by their order (e.g., “GFP-Sna3KallR” indicates an amino-terminally tagged form of Sna3 in which all lysines have been mutated to arginines). GFP localization within the vacuole lumen indicates efficient entry into the MVB pathway. Scale bar, 5 μ. (B) Plasmids containing HA-ubiquitin or GFP-tagged forms of Sna3 were transformed into _sna3_Δ _pep12_Δ cells, and lysates were generated and immunoprecipitated with anti-Sna3 antibody, followed by SDS-PAGE and Western blotting with either anti-GFP or anti-HA. (C) Pulse-chase immunoprecipitation was performed on a variety of GFP-tagged forms of Sna3, as well as endogenous Sna3 using anti-Sna3 polyclonal antisera. Degradation was quantitated using a phosphoimager, and percent remaining relative to time zero was plotted over time. The vacuolar protease deleted strain TVY614 was used to assess vacuolar degradation of endogenous Sna3. Experiments were performed at least three times to allow statistical analyses.
Figure 2.
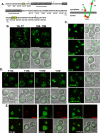
Identification of _cis_-acting sorting determinants in Sna3. (A) Sequence of Sna3 is presented to illustrate the scanning alanine mutagenesis approach, with underlined groups of residues representing mutagenized clusters. Each of the lysine residues is highlighted with an asterisk, and mutagenesis was performed in wild-type and lysine-minus (Sna3KallR) forms of Sna3, and Sna3KallR data are presented. Transmembrane domains are indicated by gray boxes, and motifs that play a role in Sna3 MVB sorting are indicated by yellow boxes. A diagram of Sna3 topology illustrates the locations of the lysine residues with red stars. (B) Residues 14-17 and 104-108 are critical for sorting of Sna3KallR into the MVB pathway. Fluorescence microscopy was used to analyze the mutants generated by scanning alanine mutagenesis. These two mutants displayed a high degree of signal at the limiting membrane of the vacuole indicating a failure to enter the MVB pathway. Scale bar, 5 μ. (C) Mutation of residues surrounding Y15 do not negatively impact Sna3KallR sorting into the MVB pathway to the degree that Y15A does. Site-directed mutagenesis was used to generate the indicated mutant forms of Sna3KallR-GFP, and fluorescence microscopy was used to visualize their localization in _sna3_Δ cells. Scale bar, 5 μ. (D) The presence of an aromatic residue at position 15 in Sna3KallR-GFP is important for delivery into the MVB pathway. Site-directed mutagenesis was used to generate the indicated mutant forms of Sna3KallR-GFP, and fluorescence microscopy was used to visualize their localization in _sna3_Δ cells. Scale bar, 5 μ. (E) Sna3 mutants localize to PtdIns(3)P-positive endosomes in addition to the vacuole. Sna3KalR-GFP, Sna3KallR14AYAA17-GFP, and Sna3KallR12AAAYAA17-GFP were coexpressed with DsRed-FYVE in wild-type cells and visualized by fluorescence microscopy. Scale bar, 5 μ.
Figure 3.
Sna3 sorting determinants and ubiquitination. (A) Sna3 sorting determinants are sufficient to confer MVB sorting. Chimeras were generated between the amino- and carboxyl-terminal sorting determinants of Sna3 and the non-MVB cargo GFP-DPAP B and analyzed by fluorescence microscopy. Scale bar, 5 μ. (B) _pep12_Δ cells were transformed with plasmids expressing the indicated GFP-tagged protein, as well as HA-ubiquitin, and used to perform immunoprecipitations with anti-GFP antibody. Immunoprecipitated material was subjected to SDS-PAGE and Western blotting with either anti-GFP, to visualize the indicated protein or anti-HA to visualize ubiquitinated forms. (C) GFP-tagged Sna3 mutants were visualized in _sna3_Δ cells using fluorescent microscopy. Scale bar, 5 μ. (D) Sna3-GFP mutants and HA-tagged ubiquitin were expressed in _sna3_Δ _pep12_Δ cells and used to make lysates that were subjected to immunoprecipitation with anti-Sna3 antisera. Immunoprecipitated material was subjected to SDS-PAGE and Western blotting with either anti-GFP or anti-HA.
Figure 4.
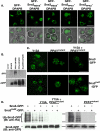
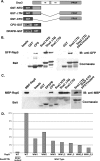
Sna3-Rsp5 interaction requires the PPAY motif within Sna3 and the WW domains of Rsp5. (A) Diagram of GST constructs used to test interactions with Rsp5. (B and C) Interaction between Sna3 Rsp5 was addressed in vitro using immobilized GST constructs and Rsp5 that was expressed in yeast (B) or bacteria (C). (D) TAP-tagged Rsp5 or WW mutant forms thereof were expressed in wild-type yeast cells and used to make lysates. Equivalent amounts of the indicated extract were incubated with GST-Sna3CTD to test the relative contributions of the WW domains. Bound material was washed, subjected to SDS-PAGE and either Coomassie stained or Western blotted with anti-actin to visualize the TAP tag, quantitated, and normalized to wild-type Rsp5.
Figure 5.
Rsp5 ligase activity is required for the sorting of Sna3KallR into the MVB pathway. (A) GFP-CPS and forms of GFP-tagged Sna3 were expressed in an rsp5G753I mutant and visualized by fluorescence microscopy. Scale bar, 5 μ. (B) Sna3-GFP and Sna3KallR-GFP were coexpressed with DsRed-FYVE in rsp5G753I cells and visualized by fluorescence microscopy. Scale bar, 5 μ. (C) Sna3-GFP and Sna3KallR-GFP were expressed in _pep12_Δ or rsp5G753I _pep12_Δ cells, and lysates were subjected to immunoprecipitation with anti-GFP antibody. Immunoprecipitated material was subjected to SDS-PAGE and Western blotting with anti-GFP or anti-ubiquitin antibodies.
Figure 6.
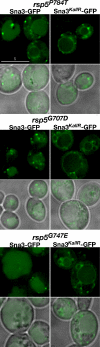
Sna3KallR-GFP is mislocalized in rsp5 HECT mutants. Sna3-GFP and Sna3KallR-GFP were expressed in rsp5P784T (MYY832), rsp5G707D (MYY833), and rsp5G747E (MYY834) cells and visualized by fluorescence microscopy. Scale bar, 5 μ.
Figure 7.
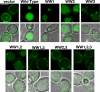
WW domains of Rsp5 are required for sorting of Sna3KallR into the MVB pathway. The rsp5G753I allele was transformed with the indicated plasmids expressing WW mutant forms of Rsp5, as well as Sna3KallR-GFP, and visualized by fluorescence microscopy. Scale bar, 5 μ.
Figure 8.
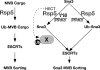
Model for the function of Rsp5 in the sorting of MVB cargoes via ubiquitin-dependent and alternative pathways. Rsp5 is responsible for the ubiquitination of a number of MVB cargoes, subsequently recognized by the ESCRTs to direct entry into the MVB pathway. Rsp5 can interact with Sna3 via WW-PPAY interactions, and this can lead to ubiquitination of Sna3 and ubiquitin-dependent recognition by ESCRTs. The interaction of Sna3 with Rsp5 appears to play an additional role in sorting via an alternate pathway that depends on Y15, presumably through the ubiquitination of an unknown sorting factor.
Similar articles
- Direct binding to Rsp5 mediates ubiquitin-independent sorting of Sna3 via the multivesicular body pathway.
McNatt MW, McKittrick I, West M, Odorizzi G. McNatt MW, et al. Mol Biol Cell. 2007 Feb;18(2):697-706. doi: 10.1091/mbc.e06-08-0663. Epub 2006 Dec 20. Mol Biol Cell. 2007. PMID: 17182850 Free PMC article. - Sna3 is an Rsp5 adaptor protein that relies on ubiquitination for its MVB sorting.
MacDonald C, Stringer DK, Piper RC. MacDonald C, et al. Traffic. 2012 Apr;13(4):586-98. doi: 10.1111/j.1600-0854.2011.01326.x. Epub 2012 Jan 31. Traffic. 2012. PMID: 22212814 Free PMC article. - Targeting of Sna3p to the endosomal pathway depends on its interaction with Rsp5p and multivesicular body sorting on its ubiquitylation.
Stawiecka-Mirota M, Pokrzywa W, Morvan J, Zoladek T, Haguenauer-Tsapis R, Urban-Grimal D, Morsomme P. Stawiecka-Mirota M, et al. Traffic. 2007 Sep;8(9):1280-96. doi: 10.1111/j.1600-0854.2007.00610.x. Epub 2007 Jul 23. Traffic. 2007. PMID: 17645729 Free PMC article. - Versatile role of the yeast ubiquitin ligase Rsp5p in intracellular trafficking.
Belgareh-Touzé N, Léon S, Erpapazoglou Z, Stawiecka-Mirota M, Urban-Grimal D, Haguenauer-Tsapis R. Belgareh-Touzé N, et al. Biochem Soc Trans. 2008 Oct;36(Pt 5):791-6. doi: 10.1042/BST0360791. Biochem Soc Trans. 2008. PMID: 18793138 Review. - The ubiquitin code of yeast permease trafficking.
Lauwers E, Erpapazoglou Z, Haguenauer-Tsapis R, André B. Lauwers E, et al. Trends Cell Biol. 2010 Apr;20(4):196-204. doi: 10.1016/j.tcb.2010.01.004. Trends Cell Biol. 2010. PMID: 20138522 Review.
Cited by
- Structural insights into the catalysis and regulation of E3 ubiquitin ligases.
Buetow L, Huang DT. Buetow L, et al. Nat Rev Mol Cell Biol. 2016 Oct;17(10):626-42. doi: 10.1038/nrm.2016.91. Epub 2016 Aug 3. Nat Rev Mol Cell Biol. 2016. PMID: 27485899 Free PMC article. Review. - The HECT domain of the ubiquitin ligase Rsp5 contributes to substrate recognition.
Lee JR, Oestreich AJ, Payne JA, Gunawan MS, Norgan AP, Katzmann DJ. Lee JR, et al. J Biol Chem. 2009 Nov 13;284(46):32126-37. doi: 10.1074/jbc.M109.048629. Epub 2009 Sep 10. J Biol Chem. 2009. PMID: 19744925 Free PMC article. - Deubiquitinating enzymes Ubp2 and Ubp15 regulate endocytosis by limiting ubiquitination and degradation of ARTs.
Ho HC, MacGurn JA, Emr SD. Ho HC, et al. Mol Biol Cell. 2017 May 1;28(9):1271-1283. doi: 10.1091/mbc.E17-01-0008. Epub 2017 Mar 15. Mol Biol Cell. 2017. PMID: 28298493 Free PMC article. - Mvb12 is a novel member of ESCRT-I involved in cargo selection by the multivesicular body pathway.
Oestreich AJ, Davies BA, Payne JA, Katzmann DJ. Oestreich AJ, et al. Mol Biol Cell. 2007 Feb;18(2):646-57. doi: 10.1091/mbc.e06-07-0601. Epub 2006 Dec 6. Mol Biol Cell. 2007. PMID: 17151358 Free PMC article. - Biogenesis and function of multivesicular bodies.
Piper RC, Katzmann DJ. Piper RC, et al. Annu Rev Cell Dev Biol. 2007;23:519-47. doi: 10.1146/annurev.cellbio.23.090506.123319. Annu Rev Cell Dev Biol. 2007. PMID: 17506697 Free PMC article. Review.
References
- Babst M., Katzmann D. J., Estepa-Sabal E. J., Meerloo T., Emr S. D. ESCRT-III: an endosome-associated heterooligomeric protein complex required for MVB sorting. Dev. Cell. 2002;3:271–282. - PubMed
- Bilodeau P. S., Urbanowski J. L., Winistorfer S. C., Piper R. C. The Vps27p Hse1p complex binds ubiquitin and mediates endosomal protein sorting. Nat. Cell Biol. 2002;4:534–539. - PubMed
- Bloom J., Amador V., Bartolini F., DeMartino G., Pagano M. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell. 2003;115:71–82. - PubMed
- Ciechanover A., Ben-Saadon R. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 2004;14:103–106. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases