Dephosphorylation of phosphotyrosine on STAT1 dimers requires extensive spatial reorientation of the monomers facilitated by the N-terminal domain - PubMed (original) (raw)
Dephosphorylation of phosphotyrosine on STAT1 dimers requires extensive spatial reorientation of the monomers facilitated by the N-terminal domain
Claudia Mertens et al. Genes Dev. 2006.
Abstract
We report experiments that infer a radical reorientation of tyrosine-phosphorylated parallel STAT1 dimers to an antiparallel form. Such a change in structure allows easy access to a phosphatase. With differentially epitope-tagged molecules, we show that the two monomers of a dimer remain together during dephosphorylation although they most likely undergo spatial reorientation. Extensive single amino acid mutagenesis within crystallographically established domains, manipulation of amino acids in an unstructured tether that connects the N-terminal domain (ND) to the core of the protein, and the demonstration that overexpressed ND can facilitate dephosphorylation of a core molecule lacking an ND all support this model: When the tyrosine-phosphorylated STAT1 disengages from DNA, the ND dimerizes and somehow assists in freeing the reciprocal pY-SH2 binding between the monomers of the dimer while ND ND dimerization persists. The core of the monomers rotate allowing reciprocal association of the coiled:coil and DNA-binding domains to present pY at the two ends of an antiparallel dimer for ready dephosphorylation.
Figures
Figure 1.
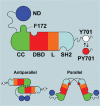
Schematic of salient features of STAT1 structure. The domains are N-terminal domain (ND), coiled:coil domain (CC), DNA-binding domain (DBD), linker domain (L), and SH2 domain (SH2). The ND is shown tied to the CC through a flexible tether (not to scale), and the residues C-terminal to the –SH2 include the Y701, which is phosphorylated (red dot) when the molecule is activated. The C-terminal region is also flexible as indicated by the wavy black line. At the bottom of the figure are diagrams of the parallel and antiparallel structures supported by crystallographic results (Chen et al. 1998; Mao et al. 2005). Notable is the F172 residue that is important in the antiparallel structure.
Figure 2.
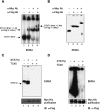
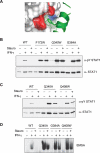
Monomers of a STAT1 phosphodimer remain associated during dephosphorylation. (A,B) Purification of STAT1-Flag:STAT1-Myc phosphodimers. 293T cells were cotransfected with expression constructs for Flag-tagged STAT1 and Myc-tagged STAT1. After IFN-γ induction, whole-cell extracts were subjected to a three-step affinity-purification scheme (see Materials and Methods). Material was evaluated by EMSA with a M67 probe before purification (A) and after purification (B) using anti-Flag and anti-Myc antibodies for shifting STAT1–DNA complexes. (C,D) Continued association of STAT1 monomers in a dimer after in vitro dephosphorylation with GST-TC45. Purified doubly tagged STAT1 phosphodimers alone (C,D, lanes 1,3) or as mixture with purified nontagged phospho STAT1β (C,D, lanes 2,4) were incubated for 60 min with purified tyrosine phosphatase TC45. (Top panels) Tyrosine phosphorylation of STAT1 proteins was monitored before and after TC45 treatment by EMSA. (Bottom panels) The presence of Flag-Myc heterodimers before and after dephosphorylation was assayed by pull-down with Ni-NTA agarose and immunoblot detection of coimmunoprecipitated proteins with anti-Flag antibodies. Note that in C approximately equal amounts of doubly tagged STAT1 (lane 2, upper band) and nontagged STAT1β (lane 2, lower band) were used, whereas in D 10-fold as much nontagged STAT1β was included to assay for a possible dissociation and reassociation of tagged dimers.
Figure 3.
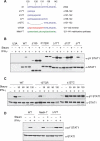
Persistent phosphorylation of STAT1 tether mutants. (A) Amino acid sequence of STAT1 tether and tether mutants. (B) Dephosphorylation of STAT1 and STAT1 mutants. U3A cells that lack STAT1 were left untransfected (U3A) or transfected for 24 h with vectors encoding wild-type STAT1 (WT), STAT1 lacking the first 135 amino acids (Δ135), or other mutant STAT1 proteins: F172W, Δ17T, Δ12T, Δ7T; as indicated in A. Parallel samples of cells were treated or not treated with IFN-γ for 30 min, and IFN-γ for 30 min followed by the tyrosine kinase inhibitor staurosporine for an additional 45 min. Total cell extracts were prepared and Western blotted with anti-pY STAT1 (top) and anti-STAT1 antibodies (bottom). (C) Time course of dephosphorylation in staurosporine-treated cells. U3A cells were complemented with wild-type STAT1 or tether mutants 12TCR (12 central amino acids reversed, shown in red in A) or Δ12TC (12 central amino acids deleted). Expressed STAT1 proteins were IFN-γ activated for 30 min followed by treatment with staurosporine for the indicated times (30, 60, 90, 120 min). Immunoblot analysis of whole-cell extracts as in B. (D) Same assay as in B with wild-type STAT1 or mutants Δ12T and MetH-T in which residues 121–141 of STAT1 are replaced with 21 amino acids of human methionine synthase (shown in green in A).
Figure 4.
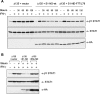
Expression of free STAT1-ND but not STAT5b-ND rescues the phenotype of mutant Δ135. (A) Coexpression of Δ135 with STAT1 ND constructs in U3A cells. Cells were transfected with indicated combinations of Δ135 and empty vector, Δ135 and HA-tagged wild-type ND, or Δ135 and HA-tagged ND-F77/L78A, which prevents ND dimerization. After IFN-γ activation followed by staurosporine treatment for indicated times (in minutes), total cell extracts were subjected to immunoblot analysis. (Bottom panel) Expression of NDs was detected with anti-HA antibodies. (B) Coexpression of Δ135 with ND of STAT1 and STAT5b. Preparation of samples as in A with staurosporine treatment for 45 min. Experiments were carried out in duplicate.
Figure 5.
Mutations of the pocket for residue F172 in the DBD cause persistent phosphorylation. (A) Shown is the crystallographically determined structure of amino acids in DBD of STAT1 that form a pocket in which the F172 residues of the CCDs interact. (B,C) STAT1 proteins with indicated mutations that were designed to disrupt the CCD/DBD dimer interface were expressed in U3A cells. After activation with IFN-γ for 30 min and staurosporine treatment for 45 min, total cell extracts were analyzed by immunoblotting with anti-pY STAT1 (top) and anti-STAT1 antibodies (bottom). (D) DNA binding of activated pocket mutants. Total cell extracts prepared as in B and C were subjected to EMSA analysis with an M67 probe.
Figure 6.
Nuclear entry of persistently phosphorylated STAT1 mutants. (A) U3A cells transfected with expression constructs for wild-type STAT1 and mutants Δ135 and Q340A were treated with IFN-γ for 30 min and staurosporine for 45 min. Whole-cell extracts (WCE) and nuclear extracts (NE) were prepared in parallel from identical samples by dividing cells evenly before lysate preparation. Samples were subjected to immunoblotting with antibodies to pY STAT1 and STAT1. (B) Flow of activated STAT1 proteins upon staurosporine addition. U3A cells expressing wild-type STAT1 or mutant proteins Δ135 and Q340A were treated with IFN-γ for 30 min followed by staurosporine treatment for the indicated times (in minutes). Cytoplasmic (Cy) and nuclear extracts (NE) were analyzed by immunoblotting.
Figure 7.
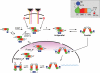
Model for STAT1 dephosphorylation using crystal structures of ND and core of STAT1. The domains and icons for STAT1 are given in Figure 1. Note that the red dot at Y701 indicates phosphorylation. First consider the parallel phosphorylated dimer (pSTAT dimer) bound to DNA. Disengagement from DNA is shown in the second panel; flexibility of the free pSTAT dimer allows contact between NDs because they are on an ∼60 Å tether (Chen et al. 1998). The ND, possibly by contacting the body of the pSTAT, induces pY–SH2 disengagement. The dimer still held together by the ND/ND interface allows rotation of the monomers so that CC/DBD interaction can occur, presenting the pY at 701 for dephosphorylation. Exit to the cytoplasm of the antiparallel nonphosphorylated dimer then occurs. In the cytoplasm, nonphosphorylated STAT1 is shown as either a monomer or dimer and can likely be tyrosine-phosphorylated in either mode. STAT4 may be obligatorily a dimer in order to be phosphorylated at the receptor (Ota et al. 2004).
Similar articles
- Clinically relevant dimer interface mutants of STAT1 transcription factor exhibit differential gene expression.
Staab J, Herrmann-Lingen C, Meyer T. Staab J, et al. PLoS One. 2013 Jul 26;8(7):e69903. doi: 10.1371/journal.pone.0069903. Print 2013. PLoS One. 2013. PMID: 23922848 Free PMC article. - Tyrosine phosphorylation regulates the partitioning of STAT1 between different dimer conformations.
Wenta N, Strauss H, Meyer S, Vinkemeier U. Wenta N, et al. Proc Natl Acad Sci U S A. 2008 Jul 8;105(27):9238-43. doi: 10.1073/pnas.0802130105. Epub 2008 Jun 30. Proc Natl Acad Sci U S A. 2008. PMID: 18591661 Free PMC article. - Structural Basis of the Inhibition of STAT1 Activity by Sendai Virus C Protein.
Oda K, Matoba Y, Irie T, Kawabata R, Fukushi M, Sugiyama M, Sakaguchi T. Oda K, et al. J Virol. 2015 Nov;89(22):11487-99. doi: 10.1128/JVI.01887-15. Epub 2015 Sep 2. J Virol. 2015. PMID: 26339056 Free PMC article. - Implications of an antiparallel dimeric structure of nonphosphorylated STAT1 for the activation-inactivation cycle.
Zhong M, Henriksen MA, Takeuchi K, Schaefer O, Liu B, ten Hoeve J, Ren Z, Mao X, Chen X, Shuai K, Darnell JE Jr. Zhong M, et al. Proc Natl Acad Sci U S A. 2005 Mar 15;102(11):3966-71. doi: 10.1073/pnas.0501063102. Epub 2005 Mar 7. Proc Natl Acad Sci U S A. 2005. PMID: 15753310 Free PMC article. - The two interfaces of the STAT1 N-terminus exhibit opposite functions in IFNγ-regulated gene expression.
Staab J, Riebeling T, Koch V, Herrmann-Lingen C, Meyer T. Staab J, et al. Mol Immunol. 2015 Oct;67(2 Pt B):596-606. doi: 10.1016/j.molimm.2015.07.015. Epub 2015 Aug 11. Mol Immunol. 2015. PMID: 26275341
Cited by
- IL-2-mTORC1 signaling coordinates the STAT1/T-bet axis to ensure Th1 cell differentiation and anti-bacterial immune response in fish.
Ai K, Li K, Jiao X, Zhang Y, Li J, Zhang Q, Wei X, Yang J. Ai K, et al. PLoS Pathog. 2022 Oct 25;18(10):e1010913. doi: 10.1371/journal.ppat.1010913. eCollection 2022 Oct. PLoS Pathog. 2022. PMID: 36282845 Free PMC article. - IκB kinase epsilon (IKK(epsilon)) regulates the balance between type I and type II interferon responses.
Ng SL, Friedman BA, Schmid S, Gertz J, Myers RM, Tenoever BR, Maniatis T. Ng SL, et al. Proc Natl Acad Sci U S A. 2011 Dec 27;108(52):21170-5. doi: 10.1073/pnas.1119137109. Epub 2011 Dec 14. Proc Natl Acad Sci U S A. 2011. PMID: 22171011 Free PMC article. - Clinically relevant dimer interface mutants of STAT1 transcription factor exhibit differential gene expression.
Staab J, Herrmann-Lingen C, Meyer T. Staab J, et al. PLoS One. 2013 Jul 26;8(7):e69903. doi: 10.1371/journal.pone.0069903. Print 2013. PLoS One. 2013. PMID: 23922848 Free PMC article. - STAT5 requires the N-domain to maintain hematopoietic stem cell repopulating function and appropriate lymphoid-myeloid lineage output.
Li G, Wang Z, Zhang Y, Kang Z, Haviernikova E, Cui Y, Hennighausen L, Moriggl R, Wang D, Tse W, Bunting KD. Li G, et al. Exp Hematol. 2007 Nov;35(11):1684-94. doi: 10.1016/j.exphem.2007.08.026. Exp Hematol. 2007. PMID: 17976521 Free PMC article. - Inducible, dose-adjustable and time-restricted reconstitution of STAT1 deficiency in vivo.
Leitner NR, Lassnig C, Rom R, Heider S, Bago-Horvath Z, Eferl R, Müller S, Kolbe T, Kenner L, Rülicke T, Strobl B, Müller M. Leitner NR, et al. PLoS One. 2014 Jan 29;9(1):e86608. doi: 10.1371/journal.pone.0086608. eCollection 2014. PLoS One. 2014. PMID: 24489749 Free PMC article.
References
- Alexander, W.S., Hilton, D.J. The role of suppressors of cytokine signaling (SOCS) proteins in regulation of the immune response. Annu. Rev. Immunol. 2004;22:503–529. - PubMed
- Bandarian, V., Pattridge, K.A., Lennon, B.W., Huddler, D.P., Matthews, R.G., Ludwig, M.L. Domain alternation switches B12-dependent methionine synthase to the activation conformation. Nat. Struct. Biol. 2001;9:53–56. - PubMed
- Becker, S., Groner, B., Muller, C.W. Three-dimensional structure of the Stat3β homodimer bound to DNA. Nature. 1998;394:145–151. - PubMed
- Braunstein, J., Brutsaert, S., Olson, R., Schindler, C. STATs dimerize in the absence of phosphorylation. J. Biol. Chem. 2003;278:34111–34140. - PubMed
- Brivanlou, A.H., Darnell, J.E., Jr. Signal transduction and the control of gene expression. Science. 2002;295:813–818. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- GM068566/GM/NIGMS NIH HHS/United States
- R01 AI032489/AI/NIAID NIH HHS/United States
- F32 GM068566/GM/NIGMS NIH HHS/United States
- AI34420/AI/NIAID NIH HHS/United States
- R37 AI034420/AI/NIAID NIH HHS/United States
- AI32489/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous