DNA methylation regulates long-range gene silencing of an X-linked homeobox gene cluster in a lineage-specific manner - PubMed (original) (raw)
DNA methylation regulates long-range gene silencing of an X-linked homeobox gene cluster in a lineage-specific manner
Masaaki Oda et al. Genes Dev. 2006.
Abstract
DNA methylation is a major epigenetic mechanism that has been suggested to control developmental gene regulation during embryogenesis, but its regulatory mechanisms remain unclear. In this report, we show that CpG islands associated with the X-linked homeobox gene cluster Rhox, which is highly expressed in the extraembryonic trophectoderm, are differentially methylated in a stage- and lineage-specific manner during the post-implantation development of mice. Inactivation of both Dnmt3a and Dnmt3b, DNA methyltransferases essential for the initiation of de novo DNA methylation, abolished the establishment of DNA methylation and the silencing of Rhox cluster genes in the embryo proper. The Dnmt3-dependent CpG-island methylation at the Rhox locus extended for a large genomic region ( approximately 1 Mb) containing the Rhox cluster and surrounding genes. Complementation experiments using embryonic stem (ES) cells deficient in the DNA methyltransferases suggested that the CpG-island methylation by Dnmt3a and Dnmt3b was restricted within this large genomic region, and did not affect the neighboring genes outside it, implicating the existence of region-specific boundaries. Our results suggest that DNA methylation plays important roles in both long-range gene silencing and lineage-specific silencing in embryogenesis.
Figures
Figure 1.
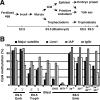
(A) A summary of cell lineages in pre- and post-implantation mouse embryogenesis. (B) Dnmt3-deficient embryos retain the global DNA methylation states that existed at the blastocyst stage. DNA methylation states of the pericentromeric major satellite repeats (black), Line1 (dark gray), IAP element (light gray), and Igf2r (white) in the embryonic tissues of male and female wild-type embryos and male mutant embryos were analyzed by bisulfite sequencing. The graph indicates the percentage of total CpG sites that were methylated in the bisulfite-sequenced clones of each region. (Emb) Embryo proper; (Troph) trophoblast; (8-cell) eight-cell embryo; (Blast) blastocyst; (ICM) inner cell mass; (WT) wild type; (DKO) _Dnmt3a_−/− _Dnmt3b_−/− (double knockout); (MT1−/−) Dnmt1−/−; (3a−/−) _Dnmt3a_−/−; (3b−/−) _Dnmt3b_−/−; (3a−/−3b+/−) _Dnmt3a_−/− Dnmt3b+/−; (3a+/−3b−/−) Dnmt3a+/− _Dnmt3b_−/−. The DNA methylation patterns of bisulfite-sequenced clones for Igf2r (DMR2) are shown in Supplementary Figure 1A.
Figure 2.
Stage- and lineage-specific DNA methylation of Rhox6 and Rhox9 by Dnmt3a and Dnmt3b. (A) Schematic diagrams of the regions around the Rhox6 and Rhox9 transcription start sites.(Top) All the exons (open boxes) of Rhox6 and Rhox9 are represented. Regions around each transcription start site (arrows) meet the criteria of a CpG island (filled gray boxes). Expanded depictions of the regions around the promoter CpG islands are shown below, indicating the CpG sites (vertical bars) and HpaII sites (asterisks). PCR primers for bisulfite sequencing (white arrowheads) covered 22 CpG sites (from −237 to +218 for the Rhox6 CpG island, and from −196 to +260 for the Rhox9 CpG island). The PCR primers for HpaII-digestion PCR (black arrowheads) covered four HpaII sites. The numbering begins with the transcription start site as +1. (B–E) DNA methylation analysis of Rhox6 and Rhox9 in the E9.5 wild-type conceptus and blastocyst (B,C) and in the E8.5 wild-type and mutant embryo proper (D,E), by bisulfite sequencing (B,D) or by HpaII-digestion PCR (C,E). (B,D) DNA methylation patterns obtained by bisulfite sequencing are shown with filled (methylated) and open (unmethylated) circles. (YS) Yolk sac. A different set of PCR primers yielded similar results (Supplementary Fig. 1D). (C,E) DNA methylation states of Rhox6 and Rhox9 assessed by HpaII-digestion PCR. Genomic DNA digested with the CpG-methylation-sensitive restriction enzyme HpaII were amplified by PCR using primers flanking the HpaII sites. (Lane —) Undigested DNA. (Lane M) CpG-methylation-insensitive MspI-digested DNA. (Lane H) HpaII-digested DNA. _Igf2r_-DMR2 served as a control for a methylated locus (Stöger et al. 1993). (F) DNA methylation analysis of the promoter region of Oct4 in trophoblast cells by HpaII-digestion PCR (Gidekel and Bergman 2002). An arbitrarily selected genomic region lacking an HpaII site on mouse chromosome 10 (no HpaII) served as a loading control.
Figure 3.
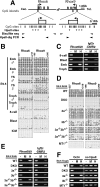
Transcriptional derepression of Rhox6 and Rhox9 in DKO embryos. (A) Expression profile of some lineage markers in DKO embryos by RT–PCR. Total RNA was isolated from the E9.5 embryo proper (Emb), yolk sac (YS), and trophoblast (Troph). (Lane Norm.1) A Dnmt3a+/− embryo. (Lane Norm.2) A Dnmt3b+/− embryo. (Lanes DKO 3 and 4) Two independent _Dnmt3a_−/− _Dnmt3b_−/− embryos. (TE) Trophectoderm; (PrEnd) primitive endoderm; (ME) mesoderm. Gapdh was used as a control for equal RNA loading. PCR cycles were shown on the right. (B) Expression profiles obtained by RT–PCR in different DNA methyltransferase knockout embryos. Total RNA was isolated from the embryo proper of an E8.5 male. Xist and IAP-type I are known to be derepressed in DKO embryos (male) and _Dnmt1_−/− embryos, respectively (Walsh et al. 1998; Sado et al. 2004). (C) The expression profiles in female and male embryos by RT–PCR. Total RNA was isolated from E8.5 male (left) and female (right) embryos.
Figure 4.
Long-range gene silencing of the Rhox gene cluster by Dnmt3a and Dnmt3b. (A) RT–PCR analysis of genes within or surrounding the Rhox cluster in mouse chromosome X A3.1 in the E9.5 conceptus of wild-type and DKO embryos. The same total RNA samples as in Figure 3A were used for this analysis. A schematic drawing of the genomic organization of this region is shown on the left (based on NCBI Build 34). Box arrows represent genes and their transcriptional directions. There is a gap of assembled mouse genome sequences within the Rhox cluster (Gap, closed box). The location of Rhox5 was mapped between Rhox3-like and Rhox6 by referring to NCBI Build 32. The vertical bold bar on the right of the RT–PCR results represents a silenced 0.7-Mb region that is transcriptionally activated in the DKO embryo proper. The RT–PCR results of Rhox6 and Rhox9 are the same data as shown in Figure 3A. Rhox7, Rhox8, Rhox10, Rhox11, and Atp1b4 were undetectable in the examined samples. (B) Section in situ hybridization of the wild-type E9.5 conceptus. Low-magnification images (left; bar is 500 μm) and high-magnification images (right; bar is 100 μm) corresponding to a region marked with a square in the low-magnification Hematoxylin and eosin (H&E) image were shown. Rhox5 and Rhox6 expressed in three layers of trophectoderm (tr): trophoblast giant cells (gi), spongiotrophoblast (sp), and labyrinthine trophoblast (la), but not in maternal decidua (de) or embryo proper (emb). Ndufa1 expressed both in trophectoderm and embryo proper. Pl1 is a marker for trophoblast giant cells. (H&E) H&E staining; (ys) yolk sac; (al) allantoic region.
Figure 5.
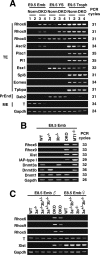
Dnmt3-dependent CpG-island methylation of genes within a large genomic region around the Rhox cluster. (A) Schematic maps of the genes used for DNA methylation analysis on mouse chromosome X A3.1. Each map represents a 2-kb genomic region around the transcription start site (arrow) with CpG islands except for Rhox5, in which a genomic region around exon 2 (a transcription start site in the epididymis) is shown. Exons are represented as open boxes. Vertical bars and asterisks indicate CpG and HpaII sites, respectively. Black and white arrowheads represent the primers for HpaII-digestion PCR or for bisulfite sequencing, respectively. (B) DNA methylation profiles of the genomic region around the Rhox cluster obtained by HpaII-digestion PCR. Genomic DNA from the blastocyst, E8.5 or E9.5 embryo proper, or trophoblast was digested with HpaII and was subjected to PCR. (Lane —) Undigested DNA. (Lane M) MspI-digested DNA. (Lane H) HpaII-digested DNA. A region lacking an HpaII site on chromosome 10 (no HpaII) served as a loading control. (8-cell) Eight-cell embryos; (Blast) blastocyst; (Emb) embryo proper; (Troph) trophoblast; (WT) wild type; (DKO) _Dnmt3a_−/− _Dnmt3b_−/−; (3a−/−) _Dnmt3a_−/−; (3b−/−) _Dnmt3b_−/−; (3a−/−3b+/−) _Dnmt3a_−/− Dnmt3b+/−; (3a+/−3b−/−) Dnmt3a+/− _Dnmt3b_−/−. (C) DNA methylation states of Ndufa1, Gm9, Rhox6, Rhox9, Cul4b, and Mcts1 in the embryo proper of wild-type and Dnmt3-mutant embryos and wild-type trophoblast by bisulfite sequencing. The graph indicates the percentage of total CpG sites that were methylated in the bisulfite-sequenced clones of each analyzed region. The results for Rhox6 and Rhox9 are based on the data shown in Figure 2. A schematic drawing of the regions around these six genes is shown below, indicating the locations and selected distances between their transcriptional start sites. Additional and detailed results are provided in Supplementary Figure 2.
Figure 6.
Dnmt3-dependent DNA methylation of the Rhox cluster genes in ES cells. Genomic DNA from wild-type (WT), _Dnmt3a_−/− (3a−/−), _Dnmt3b_−/− (3b−/−), _Dnmt3a_−/− _Dnmt3b_−/− (DKO), _Dnmt1_−/− (MT1−/−), and _Dnmt1_−/− _Dnmt3a_−/− _Dnmt3b_−/− (TKO) ES cells, and from DKO and TKO ES cells expressing the Dnmt1 (MT1), Dnmt3a (3a), Dnmt3a2 (3a2), or Dnmt3b1 (3b1) proteins were used for DNA methylation analysis. (A) DNA methylation patterns in Rhox6 and Rhox9 obtained by bisulfite sequencing are shown with filled (methylated) and open (unmethylated) circles. (B) DNA methylation profiles of the Rhox cluster genes in the ES cells obtained by HpaII-digestion PCR. (Lane —) Undigested DNA. (Lane M) MspI-digested DNA. (Lane H) HpaII-digested DNA. (C) DNA methylation states of Ndufa1, Gm9, Rhox6, Rhox9, Cul4b, and Mcts1 in ES cells. The graph indicates the percentage of total CpG sites that were methylated in bisulfite-sequenced clones of each analyzed region. Additional and detailed results are provided in Supplementary Figure 4. (D) DNA methylation patterns of Rhox6 and Rhox9 obtained by bisulfite sequencing in TKO ES cells expressing Dnmt1, Dnmt3a2, or both proteins.
Similar articles
- Ontogeny of CpG island methylation and specificity of DNMT3 methyltransferases during embryonic development in the mouse.
Auclair G, Guibert S, Bender A, Weber M. Auclair G, et al. Genome Biol. 2014;15(12):545. doi: 10.1186/s13059-014-0545-5. Genome Biol. 2014. PMID: 25476147 Free PMC article. - Dynamic transition of Dnmt3b expression in mouse pre- and early post-implantation embryos.
Hirasawa R, Sasaki H. Hirasawa R, et al. Gene Expr Patterns. 2009 Jan;9(1):27-30. doi: 10.1016/j.gep.2008.09.002. Epub 2008 Sep 11. Gene Expr Patterns. 2009. PMID: 18814855 - Preference of DNA methyltransferases for CpG islands in mouse embryonic stem cells.
Hattori N, Abe T, Hattori N, Suzuki M, Matsuyama T, Yoshida S, Li E, Shiota K. Hattori N, et al. Genome Res. 2004 Sep;14(9):1733-40. doi: 10.1101/gr.2431504. Epub 2004 Aug 12. Genome Res. 2004. PMID: 15310660 Free PMC article. - The DNMT3 family of mammalian de novo DNA methyltransferases.
Chédin F. Chédin F. Prog Mol Biol Transl Sci. 2011;101:255-85. doi: 10.1016/B978-0-12-387685-0.00007-X. Prog Mol Biol Transl Sci. 2011. PMID: 21507354 Review. - Epigenetic programming of differential gene expression in development and evolution.
Monk M. Monk M. Dev Genet. 1995;17(3):188-97. doi: 10.1002/dvg.1020170303. Dev Genet. 1995. PMID: 8565325 Review.
Cited by
- Homeobox gene Rhox5 is regulated by epigenetic mechanisms in cancer and stem cells and promotes cancer growth.
Li Q, O'Malley ME, Bartlett DL, Guo ZS. Li Q, et al. Mol Cancer. 2011 May 24;10:63. doi: 10.1186/1476-4598-10-63. Mol Cancer. 2011. PMID: 21609483 Free PMC article. - G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription.
Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. Tachibana M, et al. EMBO J. 2008 Oct 22;27(20):2681-90. doi: 10.1038/emboj.2008.192. Epub 2008 Sep 25. EMBO J. 2008. PMID: 18818694 Free PMC article. - Genome and epigenome analysis of monozygotic twins discordant for congenital heart disease.
Lyu G, Zhang C, Ling T, Liu R, Zong L, Guan Y, Huang X, Sun L, Zhang L, Li C, Nie Y, Tao W. Lyu G, et al. BMC Genomics. 2018 Jun 4;19(1):428. doi: 10.1186/s12864-018-4814-7. BMC Genomics. 2018. PMID: 29866040 Free PMC article. - Genome-wide analysis in the mouse embryo reveals the importance of DNA methylation for transcription integrity.
Dahlet T, Argüeso Lleida A, Al Adhami H, Dumas M, Bender A, Ngondo RP, Tanguy M, Vallet J, Auclair G, Bardet AF, Weber M. Dahlet T, et al. Nat Commun. 2020 Jun 19;11(1):3153. doi: 10.1038/s41467-020-16919-w. Nat Commun. 2020. PMID: 32561758 Free PMC article. - Ontogeny of CpG island methylation and specificity of DNMT3 methyltransferases during embryonic development in the mouse.
Auclair G, Guibert S, Bender A, Weber M. Auclair G, et al. Genome Biol. 2014;15(12):545. doi: 10.1186/s13059-014-0545-5. Genome Biol. 2014. PMID: 25476147 Free PMC article.
References
- Bachman, K.E., Rountree, M.R., Baylin, S.B. Dnmt3a and Dnmt3b are transcriptional repressors that exhibit unique localization properties to heterochromatin. J. Biol. Chem. 2001;276:32282–32287. - PubMed
- Bird, A. DNA methylation patterns and epigenetic memory. Genes & Dev. 2002;16:6–21. - PubMed
- Brandeis, M., Frank, D., Keshet, I., Siegfried, Z., Mendelsohn, M., Nemes, A., Temper, V., Razin, A., Cedar, H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. - PubMed
- Carninci, P., Kasukawa, T., Katayama, S., Gough, J., Frith, M.C., Maeda, N., Oyama, R., Ravasi, T., Lenhard, B., Wells, C., et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous