Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts - PubMed (original) (raw)
Aurora-A acts as a tumor suppressor and regulates self-renewal of Drosophila neuroblasts
Hongyan Wang et al. Genes Dev. 2006.
Abstract
The choice of self-renewal versus differentiation is a fundamental issue in stem cell and cancer biology. Neural progenitors of the Drosophila post-embryonic brain, larval neuroblasts (NBs), divide asymmetrically in a stem cell-like fashion to generate a self-renewing NB and a Ganglion Mother Cell (GMC), which divides terminally to produce two differentiating neuronal/glial daughters. Here we show that Aurora-A (AurA) acts as a tumor suppressor by suppressing NB self-renewal and promoting neuronal differentiation. In aurA loss-of-function mutants, supernumerary NBs are produced at the expense of neurons. AurA suppresses tumor formation by asymmetrically localizing atypical protein kinase C (aPKC), an NB proliferation factor. Numb, which also acts as a tumor suppressor in larval brains, is a major downstream target of AurA and aPKC. Notch activity is up-regulated in aurA and numb larval brains, and Notch signaling is necessary and sufficient to promote NB self-renewal and suppress differentiation in larval brains. Our data suggest that AurA, aPKC, Numb, and Notch function in a pathway that involved a series of negative genetic interactions. We have identified a novel mechanism for controlling the balance between self-renewal and neuronal differentiation during the asymmetric division of Drosophila larval NBs.
Figures
Figure 1.
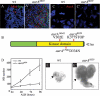
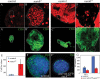
AurA is a novel tumor suppressor gene. (_A,A_′) Wild-type (A) and aurA8839 (A′) larval central brain regions at 96 h ALH were stained with the NB markers Mira (red) and Insc (green); DNA staining is in blue in all panels. (B) Schematic representation of AurA domains and mutated sites for aurA8839, aurA14641, and aurA17961. (_C,C_′) AurA is undetectable in aurA8839 mutant larval NBs. AurA in wild-type metaphase NBs (C) is localized to both centrosomes, but is undetectable in aurA8839 metaphase NBs (_C_′). (D) Quantification of wild-type and aurA8839 central brain NB numbers from 24- to 96-h-ALH. n = 20 per time point per genotype. (_E,E_′) aur8839 larval brain (_E_′) can grow to a massive size compared with wild-type brains (E). Arrows point to the brain lobes.
Figure 2.
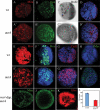
AurA acts to suppress self-renewal and promote neuronal differentiation. (_A–H_′), Confocal single-scanning images of wild-type (A–G) and aurA mutant (_A_′–_G_′) larval brains at 96 h ALH stained with NB markers Mira (_A,A_′, in red), Dpn (_B,B_′, in green), BrdU incorporation and labeling (_C,C_′, in black), phospho-Histone H3 (_D,D_′, in red), neuronal marker Elav (_E,E_′, in red), nuclear Pros (_F,F_′, in green), and CycE (_G,G_′, in red). (_H,H_′), The number of dMyc-expressing cells is increased in aurA mutant (_H_′, in green) compared with wild type (H, in green). (I–L) NB overgrowth is dependent on CDK2/CycE activity. UAS-dap; _aurA_8839 (data not shown) and wor-Gal4, UAS-dap (wor > dap); _aurA_8839 brains at 96 h ALH were stained with Mira (I, in green), Dpn (J, in green), and Elav (K, in red). (L) Quantification of NB numbers in UAS-dap; _aurA_8839 (control) and wor > dap; _aurA_8839 brains at 96 h ALH. n = 20 for each genotype. In panels that contain both central brain and optic lobe, the approximate margin between the two regions is marked by a dashed line with the central brain region to the left and optic lobe to the right (optic lobe NBs are smaller than central brain NBs). Note that in aurA mutants, both regions are enlarged compared with wild type. Posterior brain regions are imaged in all panels.
Figure 3.
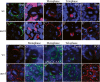
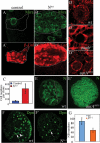
AurA is required for asymmetric localization of aPKC and Numb/Pon and regulates spindle alignment. Brat (_A,A_′), Pros (_B,B_′), and Mira (_C,C_′), remain asymmetrically localized in both wild-type (A–C) and aurA8839 (A_′–_C ′) mutant metaphase NBs; DNA staining is in blue in all panels. Mira is exclusively segregated to the GMC at telophase in both wild-type (D) and aurA mutant (_D_′) NBs. (_E,E_′) Baz has a slight expansion to the cortex but remains asymmetrically localized in aurA8839 (_E_′) compared with wild-type (E) NBs. (_F,F_′) aPKC is delocalized to the entire cortex in aurA8839 NBs (_F_′) in contrast to a crescent seen in a wild-type NB (F). (_G–G_″) During metaphase, Numb is either seen as weakly scattered to the entire cortex (_G_′) or a strongly reduced crescent (_G_″) in NBs from 120-h-ALH larvae. Numb is delocalized to the entire cortex in 24.7% of NBs and Numb crescent is reduced in 28.8% NBs from 96-h-ALH larvae (n = 73). (H) Ectopic cortical localization of aPKC leads to delocalization of Numb. When aPKC-CAAX is overexpressed, Numb is delocalized in metaphase NBs. At telophase, Numb asymmetric segregation is defective in _aurA_8839 larval NBs (_I_′) compared with that in wild-type larval NBs (I). Pon (_J_′) is also often delocalized to the entire cortex in _aurA_8839 larval NBs, while they are seen as crescents in wild-type NBs (J).
Figure 4.
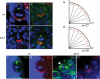
AurA controls mitotic spindle orientation through regulating asymmetric localization of Mud. (_A,A_′) The mitotic spindle is often misoriented relative to the Mira crescent in aurA17961 mutants (_A_′), in contrast to wild type (A) in which the spindle orientation inferred by two CNN-labeled centrosomes is always aligned perpendicular to the Mira crescent. CNN staining is often lost (_A_″) or strongly reduced (_A_′) in aurA8839 mutant NBs. In aurA mutants, one (data not shown) or three (_A_′″) centrosomes can also be observed. The quantification of the distribution of spindle orientation in wild-type (B) and aurA17961 mutant (_B_′) NBs based on NBs that have two CNN+ centrosomes. (_C–D_′) AurA is required for Mud asymmetric cortical localization but not its centrosomal localization. (C) In wild-type metaphase NBs, Mud is asymmetrically localized as a cortical crescent at the apical cortex as well as to both centrosomes. (_C_′) In contrast, in aurA metaphase NBs, Mud is delocalized to almost the entire cortex but remains localized to both centrosomes (arrow). (D,D′) Mira is asymmetrically localized in both wild-type NBs (D) and aurA mutant (_D_′).
Figure 5.
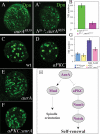
AurA acts upstream of Numb to suppress tumor formation. (A–E) Numb acts as a tumor suppressor during larval brain development. (_A_′,_B_′) NB MARCM clones marked by CD8-GFP in third instar larval brains. Most cells in numb15 clones express Dpn (B) but very few cells express Elav (D), whereas in wild-type clones usually only a single cell (the NB) expresses Dpn (A) and most cells express Elav (C). (E) Quantification of the number of cells per clone in wild-type and numb15 clones. (F–G) Overexpression of Numb (nb)-GFP suppresses the NB overproliferation phenotype in aurA8839 brains. Overexpression of Numb-GFP using wor-Gal4 in aurA8839 mutant background (wor > nb-GFP;aurA8839) reduces brain size (_F_′) compared with the control wor-Gal4, aurA8839 (F) at 120 h ALH, and the NB number is also significantly reduced (G; n = 20 per genotype per time point).
Figure 6.
Notch activity is up-regulated in aurA mutants and Notch is necessary and sufficient for NB self-renewal. (_A–B_′) Overexpression of activated Notch (the intracellular domain of Notch, Nact) induced NB overgrowth and tumor formation. (_A_′,_B_′) Mitotic clones were marked by β-galactosidase (β-Gal). In Nact clones, most cells express Mira (B), whereas only one cell (NB) expresses Mira in control clone (A). (C) Quantification of the number of cells per clone in control and activated Notch clones. (_D,D_″) Spdo localization in wild-type (D), numb15 (_D_′), and aurA8839 (_D_″) mutant larval NBs. (D) In wild-type NBs, Spdo is mostly observed as weak punctate structures on the cortex as well as throughout the cytoplasm. In numb15 (_D_′) and aurA8839 (_D_″) mutants, Spdo is almost exclusively localized at the cortex. (_E,E_′) Notch levels indicated by an antibody against its intracellular domain are up-regulated in aurA mutant brains (_E_′) compared with wild type (E). (_F,F_′) Notch is a critical proliferation factor for larval NBs. _Notchts-_1 third instar larval brains (68 h ALH, 29°C from larval hatching) contain fewer NBs, marked by Dpn in green (_F_′) compared with wild type (F). Note that wild-type larval NBs exhibit either nuclear (arrow) or cytosolic (arrowhead) Dpn signals, whereas most of the _Notchts-_1 larval NBs exhibit nuclear (arrow) but not cytosolic Dpn. (G) Quantification of NB numbers per brain lobe for wild type and _Notchts-_1. Genotypes are hs-FLP; actin-FRT-y+-FRT-Gal4, UAS-nlsLacZ(_A,A_′), and hs-FLP; actin-FRT-y+-FRT-Gal4, UAS-nlsLacZ/UAS-Nact (_B,B_′).
Figure 7.
Genetic interactions of AurA with Notch and aPKC. (_A,A_′) _Notchts-_1 suppresses the aurA overproliferation phenotype. Brains double mutant for _Notchts-_1; aurA8839 do not display overgrowth of NBs (_A_′) as indicated by the Dpn staining compared with aurA8839 (A) at 68 h ALH at 29°C. (B) Quantification of NB numbers per brain lobe for _Notchts-_1; aurA8839 and aurA8839 alone. (C–G) aPKC significantly suppresses NB overgrowth phenotype in aurA. Larval brains of wild-type (C), aPKC (D), aurA8839 (E), and aPKC; aurA8839 double mutant (F) were stained with Dpn and number of NBs per brain lobe were quantified at 68 h ALH (G). n = 20. (H) A model showing a pathway composed of AurA, aPKC, Numb, and Notch in the regulation of larval NB self-renewal. AurA is also involved in spindle orientation through regulating Mud asymmetric localization. However, in our model the proposed role for aPKC is formal and our genetic analysis does not imply a direct molecular role of AurA on aPKC.
Similar articles
- Drosophila Aurora-A kinase inhibits neuroblast self-renewal by regulating aPKC/Numb cortical polarity and spindle orientation.
Lee CY, Andersen RO, Cabernard C, Manning L, Tran KD, Lanskey MJ, Bashirullah A, Doe CQ. Lee CY, et al. Genes Dev. 2006 Dec 15;20(24):3464-74. doi: 10.1101/gad.1489406. Genes Dev. 2006. PMID: 17182871 Free PMC article. - Protein phosphatase 2A regulates self-renewal of Drosophila neural stem cells.
Wang C, Chang KC, Somers G, Virshup D, Ang BT, Tang C, Yu F, Wang H. Wang C, et al. Development. 2009 Jul;136(13):2287-96. doi: 10.1242/dev.035758. Development. 2009. PMID: 19502489 - Polo inhibits progenitor self-renewal and regulates Numb asymmetry by phosphorylating Pon.
Wang H, Ouyang Y, Somers WG, Chia W, Lu B. Wang H, et al. Nature. 2007 Sep 6;449(7158):96-100. doi: 10.1038/nature06056. Nature. 2007. PMID: 17805297 Free PMC article. - [Molecular mechanisms for asymmetric cell division in Drosophila neural stem cells].
Nishimura T. Nishimura T. Tanpakushitsu Kakusan Koso. 2009 Sep;54(11):1363-9. Tanpakushitsu Kakusan Koso. 2009. PMID: 19764471 Review. Japanese. No abstract available. - Generating neuronal diversity in the Drosophila central nervous system: a view from the ganglion mother cells.
Karcavich RE. Karcavich RE. Dev Dyn. 2005 Mar;232(3):609-16. doi: 10.1002/dvdy.20273. Dev Dyn. 2005. PMID: 15704126 Review.
Cited by
- Aurora A kinase regulates proper spindle positioning in C. elegans and in human cells.
Kotak S, Afshar K, Busso C, Gönczy P. Kotak S, et al. J Cell Sci. 2016 Aug 1;129(15):3015-25. doi: 10.1242/jcs.184416. Epub 2016 Jun 22. J Cell Sci. 2016. PMID: 27335426 Free PMC article. - Parafibromin governs cell polarity and centrosome assembly in Drosophila neural stem cells.
Deng Q, Wang C, Koe CT, Heinen JP, Tan YS, Li S, Gonzalez C, Sung WK, Wang H. Deng Q, et al. PLoS Biol. 2022 Oct 12;20(10):e3001834. doi: 10.1371/journal.pbio.3001834. eCollection 2022 Oct. PLoS Biol. 2022. PMID: 36223339 Free PMC article. - Protein phosphatase 4 mediates localization of the Miranda complex during Drosophila neuroblast asymmetric divisions.
Sousa-Nunes R, Chia W, Somers WG. Sousa-Nunes R, et al. Genes Dev. 2009 Feb 1;23(3):359-72. doi: 10.1101/gad.1723609. Genes Dev. 2009. PMID: 19204120 Free PMC article. - Drosophila neuroblast asymmetric divisions: cell cycle regulators, asymmetric protein localization, and tumorigenesis.
Chia W, Somers WG, Wang H. Chia W, et al. J Cell Biol. 2008 Jan 28;180(2):267-72. doi: 10.1083/jcb.200708159. Epub 2008 Jan 21. J Cell Biol. 2008. PMID: 18209103 Free PMC article. Review. - Mei-P26 regulates microRNAs and cell growth in the Drosophila ovarian stem cell lineage.
Neumüller RA, Betschinger J, Fischer A, Bushati N, Poernbacher I, Mechtler K, Cohen SM, Knoblich JA. Neumüller RA, et al. Nature. 2008 Jul 10;454(7201):241-5. doi: 10.1038/nature07014. Epub 2008 Jun 4. Nature. 2008. PMID: 18528333 Free PMC article.
References
- Almeida, M.S., Bray, S.J. Regulation of post-embryonic neuroblasts by Drosophila Grainyhead. Mech. Dev. 2005;122:1282–1293. - PubMed
- Androutsellis-Theotokis, A., Leker, R.R., Soldner, F., Hoeppner, D.J., Ravin, R., Poser, S.W., Rueger, M.A., Bae, S.K., Kittappa, R., McKay, R.D. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 2006;442:823–826. - PubMed
- Badano, J.L., Teslovich, T.M., Katsanis, N. The centrosome in human genetic disease. Nat. Rev. Genet. 2005;6:194–205. - PubMed
- Bello, B., Reichert, H., Hirth, F. The brain tumor gene negatively regulates neural progenitor cell proliferation in the larval central brain of Drosophila. Development. 2006;133:2639–2648. - PubMed
- Berdnik, D., Knoblich, J.A. Drosophila Aurora-A is required for centrosome maturation and actin-dependent asymmetric protein localization during mitosis. Curr. Biol. 2002;12:640–647. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous