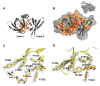Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair - PubMed (original) (raw)
Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair
Maria Victoria Botuyan et al. Cell. 2006.
Abstract
Histone lysine methylation has been linked to the recruitment of mammalian DNA repair factor 53BP1 and putative fission yeast homolog Crb2 to DNA double-strand breaks (DSBs), but how histone recognition is achieved has not been established. Here we demonstrate that this link occurs through direct binding of 53BP1 and Crb2 to histone H4. Using X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy, we show that, despite low amino acid sequence conservation, both 53BP1 and Crb2 contain tandem tudor domains that interact with histone H4 specifically dimethylated at Lys20 (H4-K20me2). The structure of 53BP1/H4-K20me2 complex uncovers a unique five-residue 53BP1 binding cage, remarkably conserved in the structure of Crb2, that best accommodates a dimethyllysine but excludes a trimethyllysine, thus explaining the methylation state-specific recognition of H4-K20. This study reveals an evolutionarily conserved molecular mechanism of targeting DNA repair proteins to DSBs by direct recognition of H4-K20me2.
Figures
Figure 1. Methylation State-Specific Recognition of Histone H4-K20 by 53BP1 Tandem Tudor Domains
(A) Representative ITC results for the titration of 53BP1 tandem tudor domains with, from left to right panels, nonmethylated, monomethylated, dimethylated, and trimethylated H4-K20 (residues 12–25). The number of stars (★) indicates the number of methyl groups attached to Nζ of Lys20. Shown for each experiment are the integrated heat measurements from raw titration data as well as curve fitting with a standard one-site model. Inset in the last panel is a control titration of JMJD2A hybrid tudor domains with trimethylated H4-K20. (B) ITC titration of 53BP1 with dimethylated H3-K79 (residues 74–83). (C) Effects of point mutations in 53BP1 tandem tudor domains on the interaction with H4-K20me2. For each interaction, the dissociation constant (KD) derived by fitting a standard one-binding site model (Wiseman et al., 1989) is reported with the associated error determined by nonlinear least squares analysis.
Figure 2. 3D Structures and Interaction of Human 53BP1 Tandem Tudor Domains
(A) Ribbon representation of the tandem tudor domains of 53BP1 in complex with dimethylated H4-K20 peptide (red stick representation). The side chains of Trp1495, Tyr1502, Phe1519, Tyr1523, and Asp1521, forming the dimethyllysine-binding cage, are shown in orange. The dimethyllysine is labeled K20me2. (B) Molecular surface representation of 53BP1 tandem tudor domains in complex with dimethylated H4-K20. 53BP1 residues with NMR weighted average chemical shift differences >0.04 ppm between free and H4-K20me2-bound 53BP1 forms are in orange. 53BP1 residues for which resonances disappeared upon peptide binding are in yellow. The H4-K20me2 peptide is in red. Inset is the view after a 180° rotation along the vertical axis. (C and D) Close-up view of the peptide-binding site in 53BP1/H4-K20me2 complex (C) and in free 53BP1 (D). The Fo – Fc and 2Fo – Fc electron density maps are overlaid for both structures (contoured at 1 σ level, blue and red mesh). Note the negative densities (red) for Trp1495 and Tyr1523 in the free protein, indicative of conformational disorder (D). Two alternative conformations are shown for the side chain of Asp1521 (D).
Figure 3. 53BP1 Uses the Same Binding Site for H4-K20me2 and H3-K79me2
(A and B) (A) 1H-15N HSQC spectra of H4-K20me2-bound (red) versus free (black) 53BP1 and (B) H3-K79me2-bound (red) versus free (black) 53BP1. H4-K20me2 (residues 16–25) and H3-K79me2 (residues 74–83) peptides were nonlabeled, while 53BP1 was 15N labeled. Peptides were added to the protein at peptide:protein ratios of 1.2:1 in (A) and 10:1 in (B). Representative 53BP1 residues perturbed (shifted or disappeared peaks) by addition of H4 and H3 peptides are labeled.
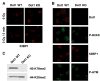
Figure 4. Lack of Histone H3 Methylation at Lys79 in Dot1-Deficient Cells Does Not Affect 53BP1 Localization to the Sites of DNA DSBs
(A) Wild-type Dot1 (Dot1 WT) and Dot1 null (Dot1 KO) primary MEFs were mock treated or irradiated with 3 Gy of IR and analyzed 5 min later by immunostaining using anti-53BP1 (53BP1) antibody. (B) Wild-type Dot1 (Dot1 WT) and Dot1 null (Dot1 KO) primary MEFs were irradiated with 1 Gy of IR and analyzed 1 hr later by immunostaining using anti-Dot1 (Dot1), anti-phosphorylated-H2AX (Ser139) (P-H2AX), anti-53BP1 (53BP1), and anti-phosphorylated-ATM kinase (Ser1981) (P-ATM) antibodies. (C) Methylation of Lys79 of histone H3 in Dot1 WT and Dot1 KO MEFs was analyzed by anti-dimethylated H3-K79 (H3-K79me2) immunoblotting. Anti-dimethylated H4-K20 (H4-K20me2) immunoblotting was used as loading control.
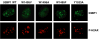
Figure 5. 53BP1 Localization to Sites of DNA DSBs Is Mediated by the Interaction between 53BP1 Tandem Tudor Domains and Dimethylated Lys20 of Histone H4
53BP1 relocalization (53BP1) in irradiated 53BP1 null MEFs transiently transfected with 53BP1 wild-type (WT) or 53BP1 mutated in the H4-K20me2-binding cage. Sites of DNA DSBs were marked by costaining using anti-phosphorylated-H2AX (Ser139) (P-H2AX) antibody.
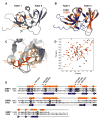
Figure 6. Crb2 Contains Tandem Tudor Domains and Interacts with Dimethylated Lys20 of Histone H4 Like 53BP1
(A) Ribbon representation of the Crb2 structure. The side chains of Phe370, Tyr378, Phe400, Thr404, and Asp402, forming the dimethyllysine-binding cage, are shown in orange. (B) Superposition of the 3D structures of 53BP1 (orange) and Crb2 (blue) in ribbon representation. (C) Magnified view of the superimposed dimethyllysine- and Arg19-binding pocket of Crb2 (brown) and 53BP1 (orange) showing key interacting residues: Phe370, Phe376, Tyr378, Phe400, Thr404, and Asp402 of Crb2; Trp1495, Tyr1500, Tyr1502, Phe1519, Tyr1523, and Asp1521 of 53BP1; and Arg19 and Lys20 (K20me2) of histone H4 peptide (red). (D) 1H-15N HSQC spectrum of 15N-labeled Crb2, free (black) and in complex with H4-K20me2 (red), showing chemical shift perturbation of a number of residues. (E) Alignment of 53BP1 and Crb2 amino acids based on their 3D structures. Red and black stars indicate 53BP1 residues interacting with K20me2 and Arg19 of H4-K20me2, respectively. In parentheses are corresponding Crb2 residues.
Similar articles
- Methylated lysine 79 of histone H3 targets 53BP1 to DNA double-strand breaks.
Huyen Y, Zgheib O, Ditullio RA Jr, Gorgoulis VG, Zacharatos P, Petty TJ, Sheston EA, Mellert HS, Stavridi ES, Halazonetis TD. Huyen Y, et al. Nature. 2004 Nov 18;432(7015):406-11. doi: 10.1038/nature03114. Epub 2004 Nov 3. Nature. 2004. PMID: 15525939 - The checkpoint Saccharomyces cerevisiae Rad9 protein contains a tandem tudor domain that recognizes DNA.
Lancelot N, Charier G, Couprie J, Duband-Goulet I, Alpha-Bazin B, Quémeneur E, Ma E, Marsolier-Kergoat MC, Ropars V, Charbonnier JB, Miron S, Craescu CT, Callebaut I, Gilquin B, Zinn-Justin S. Lancelot N, et al. Nucleic Acids Res. 2007;35(17):5898-912. doi: 10.1093/nar/gkm607. Epub 2007 Aug 28. Nucleic Acids Res. 2007. PMID: 17726056 Free PMC article. - The structural basis of modified nucleosome recognition by 53BP1.
Wilson MD, Benlekbir S, Fradet-Turcotte A, Sherker A, Julien JP, McEwan A, Noordermeer SM, Sicheri F, Rubinstein JL, Durocher D. Wilson MD, et al. Nature. 2016 Aug 4;536(7614):100-3. doi: 10.1038/nature18951. Epub 2016 Jul 27. Nature. 2016. PMID: 27462807 - Effector proteins for methylated histones: an expanding family.
Daniel JA, Pray-Grant MG, Grant PA. Daniel JA, et al. Cell Cycle. 2005 Jul;4(7):919-26. doi: 10.4161/cc.4.7.1824. Epub 2005 Jul 5. Cell Cycle. 2005. PMID: 15970672 Review. - The tale of a tail: histone H4 acetylation and the repair of DNA breaks.
Dhar S, Gursoy-Yuzugullu O, Parasuram R, Price BD. Dhar S, et al. Philos Trans R Soc Lond B Biol Sci. 2017 Oct 5;372(1731):20160284. doi: 10.1098/rstb.2016.0284. Philos Trans R Soc Lond B Biol Sci. 2017. PMID: 28847821 Free PMC article. Review.
Cited by
- KAP1 Deacetylation by SIRT1 Promotes Non-Homologous End-Joining Repair.
Lin YH, Yuan J, Pei H, Liu T, Ann DK, Lou Z. Lin YH, et al. PLoS One. 2015 Apr 23;10(4):e0123935. doi: 10.1371/journal.pone.0123935. eCollection 2015. PLoS One. 2015. PMID: 25905708 Free PMC article. - Role of histone deacetylase 2 in epigenetics and cellular senescence: implications in lung inflammaging and COPD.
Yao H, Rahman I. Yao H, et al. Am J Physiol Lung Cell Mol Physiol. 2012 Oct 1;303(7):L557-66. doi: 10.1152/ajplung.00175.2012. Epub 2012 Jul 27. Am J Physiol Lung Cell Mol Physiol. 2012. PMID: 22842217 Free PMC article. Review. - Double-strand break repair and mis-repair in 3D.
Zagelbaum J, Gautier J. Zagelbaum J, et al. DNA Repair (Amst). 2023 Jan;121:103430. doi: 10.1016/j.dnarep.2022.103430. Epub 2022 Nov 17. DNA Repair (Amst). 2023. PMID: 36436496 Free PMC article. - Molecular basis for H3K36me3 recognition by the Tudor domain of PHF1.
Musselman CA, Avvakumov N, Watanabe R, Abraham CG, Lalonde ME, Hong Z, Allen C, Roy S, Nuñez JK, Nickoloff J, Kulesza CA, Yasui A, Côté J, Kutateladze TG. Musselman CA, et al. Nat Struct Mol Biol. 2012 Dec;19(12):1266-72. doi: 10.1038/nsmb.2435. Epub 2012 Nov 11. Nat Struct Mol Biol. 2012. PMID: 23142980 Free PMC article. - Alterations of global histone H4K20 methylation during prostate carcinogenesis.
Behbahani TE, Kahl P, von der Gathen J, Heukamp LC, Baumann C, Gütgemann I, Walter B, Hofstädter F, Bastian PJ, von Ruecker A, Müller SC, Rogenhofer S, Ellinger J. Behbahani TE, et al. BMC Urol. 2012 Mar 13;12:5. doi: 10.1186/1471-2490-12-5. BMC Urol. 2012. PMID: 22413846 Free PMC article.
References
- Besant PG, Attwood PV. Mammalian histidine kinases. Biochim Biophys Acta. 2005;1754:281–290. - PubMed
- Charier G, Couprie J, Alpha-Bazin B, Meyer V, Qué méneur E, Guérois R, Callebaut I, Gilquin B, Zinn-Justin S. The Tudor tandem of 53BP1: a new structural motif involved in DNA and RG-rich peptide binding. Structure. 2004;12:1551–1562. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous