Alloantigen-enhanced accumulation of CCR5+ 'effector' regulatory T cells in the gravid uterus - PubMed (original) (raw)
Alloantigen-enhanced accumulation of CCR5+ 'effector' regulatory T cells in the gravid uterus
Marinos Kallikourdis et al. Proc Natl Acad Sci U S A. 2007.
Abstract
Regulatory T cells play an essential role in preventing fetal rejection by the maternal immune system. Here we show that, based on the expression of CCR5, regulatory T cells can be divided into a highly suppressive CCR5+ and a far less suppressive CCR5- subpopulation, suggesting that the former represent the effector arm of regulatory T cells. Although regulatory T cells from CCR5-/- gene deletion mutants still suppress, they are less effective mediators of maternal-fetal tolerance. The accumulation of CCR5+ regulatory T cells at this site appears to be enhanced by alloantigen. This finding is in stark contrast to the systemic expansion of regulatory T cells during pregnancy, which appears to be alloantigen-independent. The fact that CCR5+ regulatory T cells preferentially accumulate in the gravid uterus and that expression of CCR5 on regulatory T cells can be induced by activation lead us to propose that CCR5 is responsible for the accumulation of those regulatory T cells that have been activated by paternal antigens.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
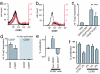
CCR5+ effector TR cells. (a) CD4+ cells were gated for the expression of CD25. CCR5 staining on total CD4+ cells (black) and on CD4+CD25+ cells (red) is shown. (b) CD4+ cells were gated for the expression of CCR5. CD25 staining on total CD4+ cells (black) and CD4+CCR5+ cells (red) is shown. (c) Relative expression of Foxp3 in CD4+ cells comparing the total population with the CD25−, CD25+CCR5−, and CD25+CCR5+ subpopulations. (d) Relative expression of Foxp3 normalized to HPRT in CD4+CCR5− and CD4+CCR5+ cells originating from CD25+ or CD25− cells by activation. (e) Effect of CCR5+ cells originating from CD25+ or CD25− on the proliferation of CD4+CD25− target cells. (f) CFSE-labeled CD4+CD25− target cells were cocultured with CD4+CD25+ CCR5+ (CCR5+) or with increasing numbers of CCR5-depleted CD4+CD25− cells (CCR5−) and activated by anti-CD3 cross-linking. The proliferation of target cells (increase in the number of CFSE-labeled cells) was analyzed by FACS on day 3.
Fig. 2.
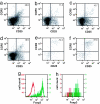
Distribution of CCR5+ TR cells in pregnant mice. Lymphocytes were isolated from allogeneically mated C57BL/6 mice at E10.5 (mid-gestation) and analyzed by FACS for the surface expression of CD4, CD25, and CCR5. Plots show CD25 and CCR5 surface expression for CD4+ gated cells from spleen (a), blood (b), iliac and lumbar lymph nodes (c), inguinal lymph nodes (d), uterus (e), and placenta (f). A representative sample is shown (n = 3). The number in the top right corner of each plot denotes the percentage of CCR5+ cells among CD4+CD25+ cells. (g and h) Intracellular Foxp3 stain of CD4+CD25+ (green) or CD4+CD25− (red) cells from spleen (g) and uterus (h) at E10.5.
Fig. 3.
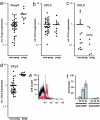
Elevated CCL4 expression in the gravid uterus and CCL4-mediated recruitment of CCR5+ TR cells. (a–d) Relative levels of Foxp3 (a), CCL3 (b), CCL5 (c), and CCL4 (d) in total uterine mRNA normalized to HPRT. Uteri were isolated from nonpregnant (non-preg.) mice or pregnant (preg.) mice at E10.5. Each point corresponds to one animal. Horizontal bars represent the mean of each data set. (e) Transwell migration assay of lymphocytes to CCL4. Input and migrated cells were analyzed by FACS for the expression of CD4 and CCR5. Histograms show CD4-gated cells. CCR5+ cells in the input cell population (black) versus the CCL4-recruited population (red). (f) Histogram comparing the migration indexes of CD4+CCR5+ and CD4+CCR5− cells recruited by CCL4 (representative experiment, n = 3), for a range of CCL4 concentrations (0, 100, 250, and 500 ng/ml). The migration index was calculated based on the number of migrated cells (_T_m), migrated cells expressing the cognate marker (_C_m), and input cells expressing the marker (_C_i).
Fig. 4.
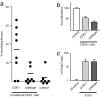
CCR5 deficiency in TR cells increases fetal loss. (a) CD25-depleted cells from C57BL/6 (H-2b) donors in combination with CD25+ cells from either CCR5−/− (H-2b) mice (CCR5−/−) or wild-type animals were injected into F5×Rag1−/− (H-2b) females. Twenty-four hours later, the mice were mated with BALB/c (H-2d) males. Allogeneic matings of wild-type animals are shown as a control. The number of intact and resorbing fetuses in all successfully mated females was scored blindly at E10.5. The percentage of resorbing over total fetuses in each animal is shown. (b) CD4+CD25− target cells were cocultured with CD25+ cells from wild-type or CCR5-deficient mice and activated with anti-CD3 cross-linking. At day 3 the proliferation of target cells was analyzed by [3H]thymidine incorporation (n = 8, representative example). (c) Relative levels of Foxp3 in total uterine RNA from wild-type or CCR5−/− pregnant mice at E10.5 normalized to HPRT. Foxp3 levels are normalized to nonpregnant wild-type mice in diestrus
Fig. 5.
The role of CCR5 in recruitment and retention of TR cells during pregnancy. Wild-type female recipients were injected with CD25+ cells prepared from CCR5-deficient mice (CCR5−/−mut., n = 4) or wild-type mice (n = 11) transduced with a retroviral vector constitutively expressing GFP. Alternatively, wild-type CD25+ cells were transduced with a retroviral vector ectopically expressing CCR5 and GFP (ect.CCR5+, n = 7). Successfully mated females were analyzed by FACS at E10.5 for the number of GFP+ cells among CD4+ cells in the uterus (a) and spleen (b). (c and d) Cells from C57BL/6 females at E10.5 of gestation after syngeneic or allogeneic mating were analyzed by FACS. Graphs show the percentage of CD4+CD25+CCR5+ cells among all cells in the uterus (c) and spleen (d). Each point corresponds to one animal. (e) Model of the recruitment and retention of TR cells in the gravid uterus.
Similar articles
- Tumor-infiltrating monocytic myeloid-derived suppressor cells mediate CCR5-dependent recruitment of regulatory T cells favoring tumor growth.
Schlecker E, Stojanovic A, Eisen C, Quack C, Falk CS, Umansky V, Cerwenka A. Schlecker E, et al. J Immunol. 2012 Dec 15;189(12):5602-11. doi: 10.4049/jimmunol.1201018. Epub 2012 Nov 14. J Immunol. 2012. PMID: 23152559 - Fetal intervention increases maternal T cell awareness of the foreign conceptus and can lead to immune-mediated fetal demise.
Wegorzewska M, Nijagal A, Wong CM, Le T, Lescano N, Tang Q, MacKenzie TC. Wegorzewska M, et al. J Immunol. 2014 Feb 15;192(4):1938-45. doi: 10.4049/jimmunol.1302403. Epub 2014 Jan 10. J Immunol. 2014. PMID: 24415782 Free PMC article. - Mechanisms of T cell tolerance towards the allogeneic fetus.
Erlebacher A. Erlebacher A. Nat Rev Immunol. 2013 Jan;13(1):23-33. doi: 10.1038/nri3361. Epub 2012 Dec 14. Nat Rev Immunol. 2013. PMID: 23237963 Review. - The role of regulatory T cells in alloantigen tolerance.
Aluvihare VR, Betz AG. Aluvihare VR, et al. Immunol Rev. 2006 Aug;212:330-43. doi: 10.1111/j.0105-2896.2006.00408.x. Immunol Rev. 2006. PMID: 16903924 Review.
Cited by
- Neutrophils in pregnancy: New insights into innate and adaptive immune regulation.
Bert S, Ward EJ, Nadkarni S. Bert S, et al. Immunology. 2021 Dec;164(4):665-676. doi: 10.1111/imm.13392. Epub 2021 Jul 29. Immunology. 2021. PMID: 34287859 Free PMC article. Review. - Regulatory T cells in nonlymphoid tissues.
Burzyn D, Benoist C, Mathis D. Burzyn D, et al. Nat Immunol. 2013 Oct;14(10):1007-13. doi: 10.1038/ni.2683. Epub 2013 Sep 18. Nat Immunol. 2013. PMID: 24048122 Free PMC article. Review. - What is the role of regulatory T cells in the success of implantation and early pregnancy?
Saito S, Shima T, Nakashima A, Shiozaki A, Ito M, Sasaki Y. Saito S, et al. J Assist Reprod Genet. 2007 Sep;24(9):379-86. doi: 10.1007/s10815-007-9140-y. J Assist Reprod Genet. 2007. PMID: 17668314 Free PMC article. Review. - Seminal fluid drives expansion of the CD4+CD25+ T regulatory cell pool and induces tolerance to paternal alloantigens in mice.
Robertson SA, Guerin LR, Bromfield JJ, Branson KM, Ahlström AC, Care AS. Robertson SA, et al. Biol Reprod. 2009 May;80(5):1036-45. doi: 10.1095/biolreprod.108.074658. Epub 2009 Jan 21. Biol Reprod. 2009. PMID: 19164169 Free PMC article. - Regulatory T-cells in pregnancy: historical perspective, state of the art, and burning questions.
Ruocco MG, Chaouat G, Florez L, Bensussan A, Klatzmann D. Ruocco MG, et al. Front Immunol. 2014 Aug 21;5:389. doi: 10.3389/fimmu.2014.00389. eCollection 2014. Front Immunol. 2014. PMID: 25191324 Free PMC article. Review.
References
- Maloy KJ, Powrie F. Nat Immunol. 2001;2:816–822. - PubMed
- Sebastiani S, Allavena P, Albanesi C, Nasorri F, Bianchi G, Traidl C, Sozzani S, Girolomoni G, Cavani A. J Immunol. 2001;166:996–1002. - PubMed
- Sakaguchi S, Sakaguchi N, Asano M, Itoh M, Toda M. J Immunol. 1995;155:1151–1164. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases