Increased nucleotide diversity with transient Y linkage in Drosophila americana - PubMed (original) (raw)
Increased nucleotide diversity with transient Y linkage in Drosophila americana
Bryant F McAllister et al. PLoS One. 2006.
Abstract
Recombination shapes nucleotide variation within genomes. Patterns are thought to arise from the local recombination landscape, influencing the degree to which neutral variation experiences hitchhiking with selected variation. This study examines DNA polymorphism along Chromosome 4 (element B) of Drosophila americana to identify effects of hitchhiking arising as a consequence of Y-linked transmission. A centromeric fusion between the X and 4(th) chromosomes segregates in natural populations of D. americana. Frequency of the X-4 fusion exhibits a strong positive correlation with latitude, which has explicit consequences for unfused 4(th) chromosomes. Unfused Chromosome 4 exists as a non-recombining Y chromosome or as an autosome proportional to the frequency of the X-4 fusion. Furthermore, Y linkage along the unfused 4 is disrupted as a function of the rate of recombination with the centromere. Inter-population and intra-chromosomal patterns of nucleotide diversity were assayed using six regions distributed along unfused 4(th) chromosomes derived from populations with different frequencies of the X-4 fusion. No difference in overall level of nucleotide diversity was detected among populations, yet variation along the chromosome exhibits a distinct pattern in relation to the X-4 fusion. Sequence diversity is inflated at loci experiencing the strongest Y linkage. These findings are inconsistent with the expected reduction in nucleotide diversity resulting from hitchhiking due to background selection or selective sweeps. In contrast, excessive polymorphism is accruing in association with transient Y linkage, and furthermore, hitchhiking with sexually antagonistic alleles is potentially responsible.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
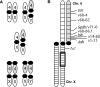
Figure 1. Characteristics of Chromosome 4 in Drosophila americana.
The unfused arrangement of Chromosome 4 is highlighted in gray. (A) Karyotypic variation involving the X and 4th chromosome as a consequence of the X-4 fusion. Frequency of the X-4 fusion determines the relative prevalence of each karyotype. Female karyotypes are expected to occur according to Hardy-Weinberg equilibrium based on a single locus with two alleles, and male karyotypes occur in proportion to the frequencies of the two arrangements of the X. (B) Idealized arrangement of Chromosome 4 during meiosis in a heterokaryotypic female having both X chromosome arrangements. Positions of the loci used in the analyses are indicated relative to the euchromatic subdivisions of Chromosome 4. Sequenced regions are underlined. An inversion of the X chromosome (In(X)c) discriminates the two lines used in the linkage analysis, and a box demarcates its approximate position.
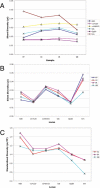
Figure 2. Nucleotide diversity on unfused Chromosome 4.
(A) Average pairwise silent diversity across populations. Samples are arranged with FP, which has the lowest frequency of the X-4 fusion, on the left and SB, which has the highest observed frequency of the X-4 fusion, on the right. No significant heterogeneity is evident among populations. (B) Pattern of sequence diversity along Chromosome 4. Loci are arranged with the centromere to the left and telomere to the right. Average pairwise silent diversity for each sample is plotted to show heterogeneity among loci and the lack of an overall pattern relative to the centromere. (C) Standardized diversity in northern populations. Total pairwise diversity in each northern sample relative to FP is plotted for each locus. A significant relationship between chromosomal position and relative diversity is indicated by the Spearman rank correlation coefficient (rs = −0.78, 95% CI: −0.94, −0.40).
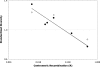
Figure 3. Relationship between mean standardized diversity in northern populations and recombination with the centromere.
Filled circles represent mean standardized diversity within the three northern samples (mean HI, IR, SB/FP) plotted in relation to estimated linkage with the centromere on a log scale. A significant (p<0.01) linear relationship is described by the plotted line, y = 0.692−0.271_x_, fitted on the log transformed recombinant fraction (R). For comparison, unfilled circles are plotted for mean diversity within the three northern samples standardized using a different southern sample [LP from ref. 24].
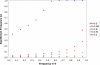
Figure 4. Sexually antagonistic alleles on Chromosome 4.
Equilibrium frequencies of a sexually antagonistic allele (A2) on the unfused arrangement are plotted for populations segregating the X-4 fusion arrangement. Loci representing a variety of different linkage relationships (R) with the centromere are illustrated. Frequency of the X-4 fusion was held constant in the simulations. Results are shown for a completely dominant antagonistic allele with a selection coefficient of 0.001 and fitness effects of opposite sign between males and females. The antagonistic allele increases male fitness and decreases female fitness.
Similar articles
- Reduced sequence variability on the Neo-Y chromosome of Drosophila americana americana.
McAllister BF, Charlesworth B. McAllister BF, et al. Genetics. 1999 Sep;153(1):221-33. doi: 10.1093/genetics/153.1.221. Genetics. 1999. PMID: 10471708 Free PMC article. - Microsatellite analysis indicates genetic differentiation of the neo-sex chromosomes in Drosophila americana americana.
Schlötterer C. Schlötterer C. Heredity (Edinb). 2000 Dec;85(Pt 6):610-6. doi: 10.1046/j.1365-2540.2000.00797.x. Heredity (Edinb). 2000. PMID: 11240628 - On the location of the gene(s) harbouring the advantageous variant that maintains the X/4 fusion of Drosophila americana.
Vieira CP, Almeida A, Dias JD, Vieira J. Vieira CP, et al. Genet Res. 2006 Jun;87(3):163-74. doi: 10.1017/S0016672306008147. Genet Res. 2006. PMID: 16817999 - How can the low levels of DNA sequence variation in regions of the drosophila genome with low recombination rates be explained?
Hudson RR. Hudson RR. Proc Natl Acad Sci U S A. 1994 Jul 19;91(15):6815-8. doi: 10.1073/pnas.91.15.6815. Proc Natl Acad Sci U S A. 1994. PMID: 8041702 Free PMC article. Review. - Adaptive hitchhiking effects on genome variability.
Andolfatto P. Andolfatto P. Curr Opin Genet Dev. 2001 Dec;11(6):635-41. doi: 10.1016/s0959-437x(00)00246-x. Curr Opin Genet Dev. 2001. PMID: 11682306 Review.
Cited by
- Drosophila americana as a model species for comparative studies on the molecular basis of phenotypic variation.
Fonseca NA, Morales-Hojas R, Reis M, Rocha H, Vieira CP, Nolte V, Schlötterer C, Vieira J. Fonseca NA, et al. Genome Biol Evol. 2013;5(4):661-79. doi: 10.1093/gbe/evt037. Genome Biol Evol. 2013. PMID: 23493635 Free PMC article. - Signals of demographic expansion in Drosophila virilis.
Mirol PM, Routtu J, Hoikkala A, Butlin RK. Mirol PM, et al. BMC Evol Biol. 2008 Feb 25;8:59. doi: 10.1186/1471-2148-8-59. BMC Evol Biol. 2008. PMID: 18298823 Free PMC article. - Local adaptation for body color in Drosophila americana.
Wittkopp PJ, Smith-Winberry G, Arnold LL, Thompson EM, Cooley AM, Yuan DC, Song Q, McAllister BF. Wittkopp PJ, et al. Heredity (Edinb). 2011 Apr;106(4):592-602. doi: 10.1038/hdy.2010.90. Epub 2010 Jul 7. Heredity (Edinb). 2011. PMID: 20606690 Free PMC article. - Simple Y-autosomal incompatibilities cause hybrid male sterility in reciprocal crosses between Drosophila virilis and D. americana.
Sweigart AL. Sweigart AL. Genetics. 2010 Mar;184(3):779-87. doi: 10.1534/genetics.109.112896. Epub 2010 Jan 4. Genetics. 2010. PMID: 20048051 Free PMC article. - Independent origins of new sex-linked chromosomes in the melanica and robusta species groups of Drosophila.
Flores SV, Evans AL, McAllister BF. Flores SV, et al. BMC Evol Biol. 2008 Jan 29;8:33. doi: 10.1186/1471-2148-8-33. BMC Evol Biol. 2008. PMID: 18230153 Free PMC article.
References
- Begun DJ, Aquadro CF. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster. Nature. 1992;356:519–520. - PubMed
- Nachman MW. Single nucleotide polymorphism and recombination rate in humans. Trends Genet. 2001;17:481–485. - PubMed
- Cutter AD, Payseur BA. Selection at linked sites in the partial selfer Caenorhabditis elegans. Mol Biol Evol. 2003;20:665–673. - PubMed
- Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8:269–294. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases