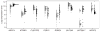Common genetic variants account for differences in gene expression among ethnic groups - PubMed (original) (raw)
Common genetic variants account for differences in gene expression among ethnic groups
Richard S Spielman et al. Nat Genet. 2007 Feb.
Abstract
Variation in DNA sequence contributes to individual differences in quantitative traits, but in humans the specific sequence variants are known for very few traits. We characterized variation in gene expression in cells from individuals belonging to three major population groups. This quantitative phenotype differs significantly between European-derived and Asian-derived populations for 1,097 of 4,197 genes tested. For the phenotypes with the strongest evidence of cis determinants, most of the variation is due to allele frequency differences at cis-linked regulators. The results show that specific genetic variation among populations contributes appreciably to differences in gene expression phenotypes. Populations differ in prevalence of many complex genetic diseases, such as diabetes and cardiovascular disease. As some of these are probably influenced by the level of gene expression, our results suggest that allele frequency differences at regulatory polymorphisms also account for some population differences in prevalence of complex diseases.
Conflict of interest statement
COMPETING INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
Figures
Figure 1
Gene expression in CHB+JPT (open circles) and CEU (filled circles) for eight genes that differ in mean expression level between the populations (N = 82 for CHB+JPT, and N = 60 for CEU). Additional information is found in Table 1.
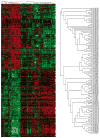
Figure 2
Results of cluster analysis. The 166 individuals are represented by columns, and the 1,097 genes of the main analysis are represented by rows. For each gene, expression level for each individual is indicated by color; intensity of red is proportional to degree of expression above the mean, and intensity of green is proportional to degree of expression below the mean. The analysis grouped the individuals into two main distinguishable groups (see enlarged tree diagram at right). One group consists of 59 CEU samples, and the other consists of the 82 CHB+JPT samples, the 24 CHLA samples and 1 CEU sample.
Figure 3
The population difference in expression of POMZP3 is accounted for by the allele frequency difference at the very closely linked SNP rs2005354. The left panel shows the distribution of expression level in the same format as in Figure 1. The right panel shows the expression level separately for individuals with each genotype of the SNP.
Comment in
- On the design and analysis of gene expression studies in human populations.
Akey JM, Biswas S, Leek JT, Storey JD. Akey JM, et al. Nat Genet. 2007 Jul;39(7):807-8; author reply 808-9. doi: 10.1038/ng0707-807. Nat Genet. 2007. PMID: 17597765 No abstract available.
Similar articles
- Interleukin-8 genetic diversity, haplotype structure and production differ in two ethnically distinct South African populations.
Paximadis M, Picton ACP, Sengupta D, Ramsay M, Puren A, Tiemessen CT. Paximadis M, et al. Cytokine. 2021 Jul;143:155489. doi: 10.1016/j.cyto.2021.155489. Epub 2021 Apr 1. Cytokine. 2021. PMID: 33814271 - Type 2 diabetes mellitus: distribution of genetic markers in Kazakh population.
Sikhayeva N, Talzhanov Y, Iskakova A, Dzharmukhanov J, Nugmanova R, Zholdybaeva E, Ramanculov E. Sikhayeva N, et al. Clin Interv Aging. 2018 Mar 5;13:377-388. doi: 10.2147/CIA.S156044. eCollection 2018. Clin Interv Aging. 2018. PMID: 29551892 Free PMC article. - Genetic architectures of ADME genes in five Eurasian admixed populations and implications for drug safety and efficacy.
Li J, Lou H, Yang X, Lu D, Li S, Jin L, Pan X, Yang W, Song M, Mamatyusupu D, Xu S. Li J, et al. J Med Genet. 2014 Sep;51(9):614-22. doi: 10.1136/jmedgenet-2014-102530. Epub 2014 Jul 29. J Med Genet. 2014. PMID: 25074363 - Predicting adult height from DNA variants in a European-Asian admixed population.
Jing X, Sun Y, Zhao W, Gao X, Ma M, Liu F, Li C. Jing X, et al. Int J Legal Med. 2019 Nov;133(6):1667-1679. doi: 10.1007/s00414-019-02039-8. Epub 2019 Apr 12. Int J Legal Med. 2019. PMID: 30976986 Retracted. - Identification and analysis of genomic regions with large between-population differentiation in humans.
Myles S, Tang K, Somel M, Green RE, Kelso J, Stoneking M. Myles S, et al. Ann Hum Genet. 2008 Jan;72(Pt 1):99-110. doi: 10.1111/j.1469-1809.2007.00390.x. Ann Hum Genet. 2008. PMID: 18184145
Cited by
- Peripheral blood gene expression as a novel genomic biomarker in complicated sarcoidosis.
Zhou T, Zhang W, Sweiss NJ, Chen ES, Moller DR, Knox KS, Ma SF, Wade MS, Noth I, Machado RF, Garcia JG. Zhou T, et al. PLoS One. 2012;7(9):e44818. doi: 10.1371/journal.pone.0044818. Epub 2012 Sep 12. PLoS One. 2012. PMID: 22984568 Free PMC article. - Variation of microRNA expression in the human placenta driven by population identity and sex of the newborn.
Guo S, Huang S, Jiang X, Hu H, Han D, Moreno CS, Fairbrother GL, Hughes DA, Stoneking M, Khaitovich P. Guo S, et al. BMC Genomics. 2021 Apr 20;22(1):286. doi: 10.1186/s12864-021-07542-0. BMC Genomics. 2021. PMID: 33879051 Free PMC article. - The general population cohort in rural south-western Uganda: a platform for communicable and non-communicable disease studies.
Asiki G, Murphy G, Nakiyingi-Miiro J, Seeley J, Nsubuga RN, Karabarinde A, Waswa L, Biraro S, Kasamba I, Pomilla C, Maher D, Young EH, Kamali A, Sandhu MS; GPC team. Asiki G, et al. Int J Epidemiol. 2013 Feb;42(1):129-41. doi: 10.1093/ije/dys234. Epub 2013 Jan 30. Int J Epidemiol. 2013. PMID: 23364209 Free PMC article. - Gene expression profiling in C57BL/6J and A/J mouse inbred strains reveals gene networks specific for brain regions independent of genetic background.
de Jong S, Fuller TF, Janson E, Strengman E, Horvath S, Kas MJ, Ophoff RA. de Jong S, et al. BMC Genomics. 2010 Jan 11;11:20. doi: 10.1186/1471-2164-11-20. BMC Genomics. 2010. PMID: 20064228 Free PMC article. - Single-nucleotide polymorphisms inside microRNA target sites influence tumor susceptibility.
Nicoloso MS, Sun H, Spizzo R, Kim H, Wickramasinghe P, Shimizu M, Wojcik SE, Ferdin J, Kunej T, Xiao L, Manoukian S, Secreto G, Ravagnani F, Wang X, Radice P, Croce CM, Davuluri RV, Calin GA. Nicoloso MS, et al. Cancer Res. 2010 Apr 1;70(7):2789-98. doi: 10.1158/0008-5472.CAN-09-3541. Epub 2010 Mar 23. Cancer Res. 2010. PMID: 20332227 Free PMC article.
References
- Schadt EE, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. - PubMed
- Cheung VG, et al. Natural variation in human gene expression assessed in lymphoblastoid cells. Nat Genet. 2003;33:422–425. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 ES015733/ES/NIEHS NIH HHS/United States
- R01 ES015733-03/ES/NIEHS NIH HHS/United States
- R01 GM081930/GM/NIGMS NIH HHS/United States
- R01 GM081930-08/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials