An acetylation site in the middle domain of Hsp90 regulates chaperone function - PubMed (original) (raw)
. 2007 Jan 12;25(1):151-9.
doi: 10.1016/j.molcel.2006.12.008.
Kenneth Robzyk, Dongxia Wang, Monica G Marcu, Shinji Tsutsumi, Kristin Beebe, Robert J Cotter, Sara Felts, David Toft, Larry Karnitz, Neal Rosen, Len Neckers
Affiliations
- PMID: 17218278
- PMCID: PMC1839984
- DOI: 10.1016/j.molcel.2006.12.008
An acetylation site in the middle domain of Hsp90 regulates chaperone function
Bradley T Scroggins et al. Mol Cell. 2007.
Abstract
Heat-shock protein 90 (Hsp90) chaperones a key subset of signaling proteins and is necessary for malignant transformation. Hsp90 is subject to an array of posttranslational modifications that affect its function, including acetylation. Histone deacetylase (HDAC) inhibitors and knockdown of HDAC6 induce Hsp90 acetylation and inhibit its activity. However, direct determination of the functional consequences of Hsp90 acetylation has awaited mapping of specific sites. We now demonstrate that Hsp90 K294 is acetylated. Mutational analysis of K294 shows that its acetylation status is a strong determinant of client protein and cochaperone binding. In yeast, Hsp90 mutants that cannot be acetylated at K294 have reduced viability and chaperone function compared to WT or to mutants that mimic constitutive acetylation. These data suggest that acetylation/deacetylation of K294 plays an important role in regulating the Hsp90 chaperone cycle.
Figures
Figure 1
Hsp90 is Acetylated in More Than One Domain (A) SkBr3 cells were labeled with [14C]acetate. Hsp90 was immunoprecipitated from cell lysate, digested with 50 (lane 2) or 100 μg/ml trypsin (lane 3), and fragments were separated by SDS-PAGE and transferred to PVDF membrane. Acetylated bands were detected by autoradiogram. The 3 indicated bands were sequenced. (B) Diagram of Hsp90 domain architecture showing location and N-terminal sequence of the bands indicated in (A). (ATPase = ATPase domain, +/-= charged linker region, Middle = middle domain, C-t = carboxy-terminal domain; the human Hsp90αsequence was used to determine amino acid numbers). K294 is identified by bolding and an asterisk. (C) Diagram of K294 relative to Hsp90 domain architecture (domains are as indicated in Fig. 1B). Below is an alignment of the sequence including and around K294 from different organisms and isoforms of Hsp90. K294 or corresponding residues in other organisms/isoforms is underlined and in bold.
Figure 2
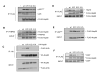
Mutation of K294 Affects Hsp90 Interaction with Client Proteins (A) COS7 cells were co-transfected with ErbB2 (+) and indicated FLAG-Hsp90 constructs. Cells were lysed and FLAG-Hsp90 immunoprecipitates were separated by SDS-PAGE; ErbB2 binding was detected by immunoblotting. (B) SkBr3 cells were transfected with indicated FLAG-Hsp90 constructs. Hsp90 was immunoprecipitated from cell lysates and ErbB2 binding was determined as in (A). (C) NIH3T3 cells stably transfected with v-src were transfected with indicated FLAG-Hsp90 constructs. P60v-Src was immunoprecipitated.and associated FLAG-Hsp90 was detected by immunoblotting. (D) SkBr3 cells were transfected with indicated FLAG-Hsp90 constructs. Endogenous mutant p53 was immunoprecipitated and associated FLAG-Hsp90 was detected as in (C). (E) COS7 cells were co-transfected with HA-HIF-1αand indicated FLAG-Hsp90 constructs. HA-HIF-1αwas immunoprecipitated and associated FLAG-Hsp90 was detected as in (C). (F) COS7 cells were co-transfected with androgen receptor and indicated FLAG-Hsp90 constructs. FLAG-Hsp90 was immunoprecipitated and androgen receptor was detected by immunoblotting.
Figure 3
Mutation of K294 Affects Hsp90 Interaction with Co-chaperones (A) COS7 cells were transfected with indicated FLAG-Hsp90 constructs. FLAG-Hsp90 was immunoprecipitated and associated p23 and p50cdc37 were detected by immunoblotting. (B) COS7 cells were transfected as in (A). FKBP52 was immunoprecipitated and Hsp90 association was detected by immunoblotting. (C) COS7 cells were co-transfected with FLAG-Aha1 and indicated HA-Hsp90 constructs. HA-Hsp90 was immunoprecipitated and associated FLAG-Aha1 was detected by immunoblotting. (D) COS7 cells were transfected with indicated FLAG-Hsp90 constructs. FLAG-Hsp90 was immunoprecipitated and Hsp70 binding was detected by immunoblotting. (E) COS7 cells were transfected with indicated FLAG-Hsp90 constructs. P60Hop was immunoprecipitated and FLAG-Hsp90 binding was detected by immunoblotting. (F) COS7 cells were co-transfected with CHIP and indicated FLAG-Hsp90 constructs. FLAG-Hsp90 was immunoprecipitated and CHIP binding was detected by immunoblotting.
Figure 4
Acetylation Status of Hsp90 K294 Affects Chk1 Kinase Activity In Vitro and Yeast Viability In Vivo (A) Purified GST-Chk1(1-265) bound to GSH-agarose was incubated with purified WT or indicated mutant Hsp90αproteins together with Hsp70, Ydj, p50cdc37, p60Hop, p23, and CK2. Reactions were washed and precipitates were incubated with [γ-32P]ATP and GST-Cdc25C substrate in kinase buffer. Kinase reaction products were separated by SDS-PAGE and transferred to PVDF membrane. Radiolabeled proteins were imaged and quantitated by phosphorimager. Fold of activation is graphed and standard errors calculated from two experiments. (B) An S. cerevisiae strain (YKR314) deleted for both genomic copies of yeast HSP90 but carrying both wild type yeast HSP82 (on a URA3 marked plasmid) and human HSP90 constructs (on a low copy CEN-TRP1 marked plasmid) was grown at 30°C to mid-log phase in liquid media. Cultures were adjusted to the same cell density before 5-fold serial dilutions were spotted onto SD-TRP plates either with or without 5-FOA and incubated at 30°C for 3 days before being photographed. (C) TRP+, ura-colonies were isolated from those strains in (B) which were viable on 5-FOA and grown at 30°C to mid-log phase in liquid YPD media. Cultures were adjusted to the same cell density before 5-fold serial dilutions were spotted onto YPD plates and incubated at 37°C for 3 days before being photographed. (E) Yeast cells transformed with a GALp-v-src-2_μ_-HIS3 construct (kindly provided by A. Caplan) and expressing the indicated human _HSP90_α constructs from CEN-ARS-TRP1 plasmids as the sole source of Hsp90 were grown in liquid SD-HIS media at 30°C to mid-log phase. Cultures were adjusted to the same cell density before serial dilutions were spotted onto either an SD-HIS plate (v-src not expressed; incubated 3 days) or an SGal-HIS plate (v-src is induced; incubated 5 days). Since p60v-Src expression is toxic to yeast, growth indicates that p60v-Src is not functionally expressed while failure to grow on galactose indicates the presence of active p60v-Src protein.
Similar articles
- Enhancement of stress resilience through histone deacetylase 6-mediated regulation of glucocorticoid receptor chaperone dynamics.
Jochems J, Teegarden SL, Chen Y, Boulden J, Challis C, Ben-Dor GA, Kim SF, Berton O. Jochems J, et al. Biol Psychiatry. 2015 Feb 15;77(4):345-55. doi: 10.1016/j.biopsych.2014.07.036. Epub 2014 Aug 28. Biol Psychiatry. 2015. PMID: 25442004 Free PMC article. - HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor.
Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. Kovacs JJ, et al. Mol Cell. 2005 May 27;18(5):601-7. doi: 10.1016/j.molcel.2005.04.021. Mol Cell. 2005. PMID: 15916966 - Uncovering a region of heat shock protein 90 important for client binding in E. coli and chaperone function in yeast.
Genest O, Reidy M, Street TO, Hoskins JR, Camberg JL, Agard DA, Masison DC, Wickner S. Genest O, et al. Mol Cell. 2013 Feb 7;49(3):464-73. doi: 10.1016/j.molcel.2012.11.017. Epub 2012 Dec 20. Mol Cell. 2013. PMID: 23260660 Free PMC article. - Posttranslational modification and beyond: interplay between histone deacetylase 6 and heat-shock protein 90.
Liu P, Xiao J, Wang Y, Song X, Huang L, Ren Z, Kitazato K, Wang Y. Liu P, et al. Mol Med. 2021 Sep 16;27(1):110. doi: 10.1186/s10020-021-00375-3. Mol Med. 2021. PMID: 34530730 Free PMC article. Review. - Drugging the HDAC6-HSP90 interplay in malignant cells.
Krämer OH, Mahboobi S, Sellmer A. Krämer OH, et al. Trends Pharmacol Sci. 2014 Oct;35(10):501-9. doi: 10.1016/j.tips.2014.08.001. Epub 2014 Sep 16. Trends Pharmacol Sci. 2014. PMID: 25234862 Review.
Cited by
- Fungal Hsp90: a biological transistor that tunes cellular outputs to thermal inputs.
Leach MD, Klipp E, Cowen LE, Brown AJ. Leach MD, et al. Nat Rev Microbiol. 2012 Oct;10(10):693-704. doi: 10.1038/nrmicro2875. Nat Rev Microbiol. 2012. PMID: 22976491 Free PMC article. Review. - Inhibiting the HSP90 chaperone destabilizes macrophage migration inhibitory factor and thereby inhibits breast tumor progression.
Schulz R, Marchenko ND, Holembowski L, Fingerle-Rowson G, Pesic M, Zender L, Dobbelstein M, Moll UM. Schulz R, et al. J Exp Med. 2012 Feb 13;209(2):275-89. doi: 10.1084/jem.20111117. Epub 2012 Jan 23. J Exp Med. 2012. PMID: 22271573 Free PMC article. - Mechanisms of Resistance to Hsp90 Inhibitor Drugs: A Complex Mosaic Emerges.
Piper PW, Millson SH. Piper PW, et al. Pharmaceuticals (Basel). 2011 Oct 25;4(11):1400-1422. doi: 10.3390/ph4111400. Pharmaceuticals (Basel). 2011. PMID: 27721330 Free PMC article. Review. - Targeting Huntington's disease through histone deacetylases.
Gray SG. Gray SG. Clin Epigenetics. 2011 Aug;2(2):257-77. doi: 10.1007/s13148-011-0025-7. Epub 2011 Feb 18. Clin Epigenetics. 2011. PMID: 22704341 Free PMC article. - Novel Promising Antifungal Target Proteins for Conquering Invasive Fungal Infections.
Zhen C, Lu H, Jiang Y. Zhen C, et al. Front Microbiol. 2022 Jun 16;13:911322. doi: 10.3389/fmicb.2022.911322. eCollection 2022. Front Microbiol. 2022. PMID: 35783432 Free PMC article. Review.
References
- Arlander SJ, Felts SJ, Wagner JM, Stensgard B, Toft DO, Karnitz LM. Chaperoning checkpoint kinase 1 (Chk1), an Hsp90 client, with purified chaperones. J Biol Chem. 2006;281:2989–2998. - PubMed
- Bali P, Pranpat M, Bradner J, Balasis M, Fiskus W, Guo F, Rocha K, Kumaraswamy S, Boyapalle S, Atadja P, et al. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J Biol Chem. 2005a;280:26729–26734. - PubMed
- Bali P, Pranpat M, Swaby R, Fiskus W, Yamaguchi H, Balasis M, Rocha K, Wang HG, Richon V, Bhalla K. Activity of suberoylanilide hydroxamic Acid against human breast cancer cells with amplification of her-2. Clin Cancer Res. 2005b;11:6382–6389. - PubMed
- Blagg BS, Kerr TD. Hsp90 inhibitors: small molecules that transform the Hsp90 protein folding machinery into a catalyst for protein degradation. Med Res Rev. 2006;26:310–338. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous