Mosaic structure of p1658/97, a 125-kilobase plasmid harboring an active amplicon with the extended-spectrum beta-lactamase gene blaSHV-5 - PubMed (original) (raw)
Mosaic structure of p1658/97, a 125-kilobase plasmid harboring an active amplicon with the extended-spectrum beta-lactamase gene blaSHV-5
M Zienkiewicz et al. Antimicrob Agents Chemother. 2007 Apr.
Abstract
Escherichia coli isolates recovered from patients during a clonal outbreak in a Warsaw, Poland, hospital in 1997 produced different levels of an extended-spectrum beta-lactamase (ESBL) of the SHV type. The beta-lactamase hyperproduction correlated with the multiplication of ESBL gene copies within a plasmid. Here, we present the complete nucleotide sequence of plasmid p1658/97 carried by the isolates recovered during the outbreak. The plasmid is 125,491 bp and shows a mosaic structure in which all modules constituting the plasmid core are homologous to those found in plasmids F and R100 and are separated by segments of homology to other known regions (plasmid R64, Providencia rettgeri genomic island R391, Vibrio cholerae STX transposon, Klebsiella pneumoniae or E. coli chromosomes). Plasmid p1658/97 bears two replication systems, IncFII and IncFIB; we demonstrated that both are active in E. coli. The presence of an active partition system (sopABC locus) and two postsegregational killing systems (pemIK and hok/sok) indicates that the plasmid should be stably maintained in E. coli populations. The conjugative transfer is ensured by the operons of the tra and trb genes. We also demonstrate that the plasmidic segment undergoing amplification contains the blaSHV-5 gene and is homologous to a 7.9-kb fragment of the K. pneumoniae chromosome. The amplicon displays the structure of a composite transposon of type I.
Figures
FIG. 1.
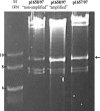
SwaI digestion of p1658/97 and p1657/97. The results of SwaI digestion of p1658/97 DNA with the “nonamplified” and the “amplified” regions isolated from E. coli colonies collected from outside and inside the growth inhibition zone (“susceptible” and “resistant,” respectively) and p1657/97 isolated from DH5α cultured with the presence of 128 μg/ml ceftazidime are shown. Equal amounts of all plasmids were loaded. The lanes are described at the top. The arrow shows the amplified fragment of 8,817 bp. Lane M, GeneRuler DNA Ladder Mix (MBI Fermentas). Numbers on the left are in base pairs.
FIG. 2.
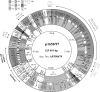
Graphical overview of p1658/97. ORFs are shown by arrows: p1658/97-specific ORFs are in black, ORFs homologous to genes of known function are in white, and ORFs homologous to genes of unknown function are in gray. The positions of mobile elements are additionally shown as white boxes. The orientations of the mobile elements are marked with black arrows; incomplete mobile elements are marked by a Δ symbol. Regions of over 90% similarity (at the nucleotide level) to the following DNA molecules are shown by shadings according to the legend: 1, plasmid F; 2, plasmid R100; 3, both plasmids F and R100; 4, the chromosomes of both E. coli and S. enterica serovar Typhimurium; 5, a region found in the K. pneumoniae chromosome and plasmids pHNM1, pSEM, and pACM1; 6, P. rettgeri genomic island R391 and the Vibrio cholerae STX transposon; 7, plasmid R64; 8, p1658/97-specific regions; 9, a region shared with pSEM, pACM1, and some other plasmids containing an integron; and 10, DNA molecules other than those mentioned above. The functional modules of p1658/97 outside the circle are indicated as follows: PAR, partition; PSK, postsegregational killing; REP, replication. The G+C contents of the sequence blocks are denoted in the inner circle.
FIG. 3.
Schematic representation of the multiresistance locus of p1658/97 with the amplicon and the integron. Black arrows indicate IS_26_, gray arrows represent genes localized within the integron and the amplicon, and white arrows indicate the remnant of IS_26_ (ΔIS_26_) and the _bla_SHV-5 gene. The position of SwaI and the sizes of the restriction fragments are indicated.
FIG. 4.
Hypothetical events leading to the present structure of p1658/97. The partial p1658/97 sequence (positions 35000 to 114500) is drawn to scale; the remaining part (including the tra region) is out of scale. The module “IS_26_cp1-spacing region-IS_26_cp4” (positions 47137 to 95371) is marked at the top. Open boxes represent the mobile elements, and arrows represent ORFs. The orientations of the mobile elements are marked with black arrows; incomplete mobile elements are marked with a Δ symbol. Dotted lines show hypothetical mobilization events that implant different submodules. The regions of the plasmid backbone are marked: REP, replication; PAR, partition; PSK, postsegregational killing. DRs, when they have been identified, are shown. Regions of over 90% similarity (at the nucleotide level) to DNA molecules other than F and R100 are shown.
Similar articles
- Tandem multiplication of the IS26-flanked amplicon with the bla(SHV-5) gene within plasmid p1658/97.
Zienkiewicz M, Kern-Zdanowicz I, Carattoli A, Gniadkowski M, Cegłowski P. Zienkiewicz M, et al. FEMS Microbiol Lett. 2013 Apr;341(1):27-36. doi: 10.1111/1574-6968.12084. Epub 2013 Feb 11. FEMS Microbiol Lett. 2013. PMID: 23330672 - Resistance to extended-spectrum beta-lactams by the emergence of SHV-12 and the loss of OmpK35 in Klebsiella pneumoniae and Escherichia coli in Malaysia.
Palasubramaniam S, Muniandy S, Navaratnam P. Palasubramaniam S, et al. J Microbiol Immunol Infect. 2009 Apr;42(2):129-33. J Microbiol Immunol Infect. 2009. PMID: 19597644 - Prevalence of acquired fosfomycin resistance among extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae clinical isolates in Korea and IS26-composite transposon surrounding fosA3.
Lee SY, Park YJ, Yu JK, Jung S, Kim Y, Jeong SH, Arakawa Y. Lee SY, et al. J Antimicrob Chemother. 2012 Dec;67(12):2843-7. doi: 10.1093/jac/dks319. Epub 2012 Aug 14. J Antimicrob Chemother. 2012. PMID: 22893681 - SHV-5 extended-spectrum beta-lactamases in clinical isolates of Escherichia coli in Malaysia.
Subramaniam G, Palasubramaniam S, Navaratnam P. Subramaniam G, et al. Indian J Med Microbiol. 2006 Jul;24(3):205-7. Indian J Med Microbiol. 2006. PMID: 16912441
Cited by
- Resistance plasmid families in Enterobacteriaceae.
Carattoli A. Carattoli A. Antimicrob Agents Chemother. 2009 Jun;53(6):2227-38. doi: 10.1128/AAC.01707-08. Epub 2009 Mar 23. Antimicrob Agents Chemother. 2009. PMID: 19307361 Free PMC article. Review. No abstract available. - Integrons: Vehicles and pathways for horizontal dissemination in bacteria.
Domingues S, da Silva GJ, Nielsen KM. Domingues S, et al. Mob Genet Elements. 2012 Sep 1;2(5):211-223. doi: 10.4161/mge.22967. Mob Genet Elements. 2012. PMID: 23550063 Free PMC article. - The resistomes of six carbapenem-resistant pathogens - a critical genotype-phenotype analysis.
Johnning A, Karami N, Tång Hallbäck E, Müller V, Nyberg L, Buongermino Pereira M, Stewart C, Ambjörnsson T, Westerlund F, Adlerberth I, Kristiansson E. Johnning A, et al. Microb Genom. 2018 Nov;4(11):e000233. doi: 10.1099/mgen.0.000233. Epub 2018 Nov 21. Microb Genom. 2018. PMID: 30461373 Free PMC article. - Sequence of conjugative plasmid pIP1206 mediating resistance to aminoglycosides by 16S rRNA methylation and to hydrophilic fluoroquinolones by efflux.
Périchon B, Bogaerts P, Lambert T, Frangeul L, Courvalin P, Galimand M. Périchon B, et al. Antimicrob Agents Chemother. 2008 Jul;52(7):2581-92. doi: 10.1128/AAC.01540-07. Epub 2008 May 5. Antimicrob Agents Chemother. 2008. PMID: 18458128 Free PMC article. - Insights into the environmental resistance gene pool from the genome sequence of the multidrug-resistant environmental isolate Escherichia coli SMS-3-5.
Fricke WF, Wright MS, Lindell AH, Harkins DM, Baker-Austin C, Ravel J, Stepanauskas R. Fricke WF, et al. J Bacteriol. 2008 Oct;190(20):6779-94. doi: 10.1128/JB.00661-08. Epub 2008 Aug 15. J Bacteriol. 2008. PMID: 18708504 Free PMC article.
References
- Blattner, F. R., I. Plunkett, G., C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases