Structural and functional characterization of the aryl hydrocarbon receptor ligand binding domain by homology modeling and mutational analysis - PubMed (original) (raw)
Structural and functional characterization of the aryl hydrocarbon receptor ligand binding domain by homology modeling and mutational analysis
Alessandro Pandini et al. Biochemistry. 2007.
Abstract
The aryl hydrocarbon receptor (AhR) is a ligand-dependent transcription factor that is activated by a structurally diverse array of synthetic and natural chemicals, including toxic halogenated aromatic hydrocarbons such as 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD). Analysis of the molecular events occurring in the AhR ligand binding and activation processes requires structural information on the AhR Per-Arnt-Sim (PAS) B-containing ligand binding domain, for which no experimentally determined structure has been reported. With the availability of extensive structural information on homologous PAS-containing proteins, a reliable model of the mouse AhR PAS B domain was developed by comparative modeling techniques. The PAS domain structures of the functionally related hypoxia-inducible factor 2alpha (HIF-2alpha) and AhR nuclear translocator (ARNT) proteins, which exhibit the highest degree of sequence identity and similarity with AhR, were chosen to develop a two-template model. To confirm the features of the modeled domain, the effects of point mutations in selected residue positions on both TCDD binding to the AhR and TCDD-dependent transformation and DNA binding were analyzed. Mutagenesis and functional analysis results are consistent with the proposed model and confirm that the cavity modeled in the interior of the domain is indeed involved in ligand binding. Moreover, the physicochemical characteristics of some residues and of their mutants, along with the effects of mutagenesis on TCDD and DNA binding, also suggest some key features that are required for ligand binding and activation of mAhR at a molecular level, thus providing a framework for further studies.
Figures
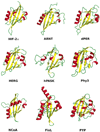
FIGURE 1
Cartoon representation of the PAS domain structures included in the analysis with their cofactors: HIF-2α (PDB ID 1P97), ARNT (PDB ID 1X0O), dPER (PDB ID 1WA9), HERG (PDB ID 1BYW), hPASK (PDB ID 1LL8), Phy3 (PDB ID 1G28), NCoA (PDB ID 1OJ5), FixL (PDB ID 1DRM), and PYP (PDB ID 1NWZ). Secondary structure attribution was according to the Kabsch and Sander method (58). For FixL and PYP the additional extra-domain elements included in the X-ray structures are also included (a long helix at the C-terminus for FixL and an N-terminal bundle of two helices for PYP).
FIGURE 2
(a) Diagram of the typical PAS fold with secondary structure elements labeled according to the nomenclature generally adopted for the PAS structures. (b) Sequence alignment of the mAhR against the templates HIF-2α and ARNT, pairwise aligned according to DALI. Only residues that are identical or similar for at least two of the three sequences are highlighted by colors. Coloring scheme for residues: red, acidic; blue, basic; purple, polar; yellow, Cys; brown, aromatic; green, hydrophobic; orange, Ser, Thr; gray, Pro, Gly. The mAhR predicted secondary structure and the template secondary structures, attributed according to the method of Kabsch and Sander (58), are also shown. Helices and β-strands are represented as white and black bars, respectively, and labeled with the PAS structure nomenclature (see panel a). Red arrows indicate the boundary residues of the mAhR cavity that have considerably smaller side chains than the corresponding ones in HIF-2α.
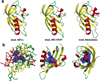
FIGURE 3
(a) Cartoon representation of the three modeled structures of the mAhR LBD. (b) Stick and cartoon representations of the model based on the HIF-2α and ARNT template structures (mod_HIF/ARNT) in different orientations, with the molecular surface (in blue) including the available volume in the cavity identified by CASTp. Secondary structure attribution was according to the method of Kabsch and Sander (58).
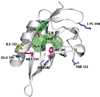
FIGURE 4
Cartoon representation of the modeled mAhR LBD (mod_HIF/ARNT) showing selected residues that were mutated. Residues with side chains pointing outside the modeled LBD are shown in blue; boundary residues of the cavity with side chains pointing inside it are shown in purple; Ile332, which is expected to have a structural role, is shown in yellow. The molecular surface (in green) including the available volume in the cavity identified by CASTp is shown.
FIGURE 5
Effect of mutation of selected residues within the mAhR LBD on TCDD-dependent AhR DNA binding. In vitro expressed wild-type or mutant AhR and wt ARNT were incubated with TCDD, and inducible AhR–ARNT–DRE complex formation was determined by gel retardation analysis as described under Materials and Methods. The positions of the induced AhR–ARNT–DRE complex are indicated by an arrow. Quantitation of the amount of the TCDD–AhR–ARNT–DRE complex was determined by phosphorimager analysis, and the results of multiple receptor preparations and gel retardation analyses (n ≥ 3) are presented in Table 4.
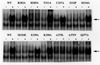
FIGURE 6
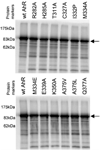
Expression levels of in vitro synthesized wild-type and mutant AhRs. 35S-Labeled wild-type and mutant AhRs were synthesized in vitro, denatured, and resolved by SDS–polyacrylamide gel electrophoresis and autoradiography of the dried gels as described in Materials and Methods. An arrow shows the bands of the AhR.
FIGURE 7
Stick representation of the helical connector main chain in the modeled mAhR LBD subjected to 1 ns MD simulation: (a) wild-type mAhR; (b) I332P mutant. The coloring scheme is according to the atom types. Hydrogen bonds are highlighted in yellow. Cartoon representation of the domain is shown in transparency.
Similar articles
- The tertiary structures of porcine AhR and ARNT proteins and molecular interactions within the TCDD/AhR/ARNT complex.
Orlowska K, Molcan T, Swigonska S, Sadowska A, Jablonska M, Nynca A, Jastrzebski JP, Ciereszko RE. Orlowska K, et al. J Mol Graph Model. 2016 Jun;67:119-26. doi: 10.1016/j.jmgm.2016.05.012. Epub 2016 May 26. J Mol Graph Model. 2016. PMID: 27288759 - Detection of the TCDD binding-fingerprint within the Ah receptor ligand binding domain by structurally driven mutagenesis and functional analysis.
Pandini A, Soshilov AA, Song Y, Zhao J, Bonati L, Denison MS. Pandini A, et al. Biochemistry. 2009 Jun 30;48(25):5972-83. doi: 10.1021/bi900259z. Biochemistry. 2009. PMID: 19456125 Free PMC article. - New aryl hydrocarbon receptor homology model targeted to improve docking reliability.
Motto I, Bordogna A, Soshilov AA, Denison MS, Bonati L. Motto I, et al. J Chem Inf Model. 2011 Nov 28;51(11):2868-81. doi: 10.1021/ci2001617. Epub 2011 Nov 2. J Chem Inf Model. 2011. PMID: 21981577 Free PMC article. - Mechanisms of ligand-induced aryl hydrocarbon receptor-mediated biochemical and toxic responses.
Wilson CL, Safe S. Wilson CL, et al. Toxicol Pathol. 1998 Sep-Oct;26(5):657-71. doi: 10.1177/019262339802600510. Toxicol Pathol. 1998. PMID: 9789953 Review. - Aryl hydrocarbon receptor-mediated signal transduction.
Rowlands JC, Gustafsson JA. Rowlands JC, et al. Crit Rev Toxicol. 1997 Mar;27(2):109-34. doi: 10.3109/10408449709021615. Crit Rev Toxicol. 1997. PMID: 9099515 Review.
Cited by
- The Cellular and Molecular Determinants of Naphthoquinone-Dependent Activation of the Aryl Hydrocarbon Receptor.
Faber SC, Giani Tagliabue S, Bonati L, Denison MS. Faber SC, et al. Int J Mol Sci. 2020 Jun 9;21(11):4111. doi: 10.3390/ijms21114111. Int J Mol Sci. 2020. PMID: 32526934 Free PMC article. - Ginsenosides are novel naturally-occurring aryl hydrocarbon receptor ligands.
Hu Q, He G, Zhao J, Soshilov A, Denison MS, Zhang A, Yin H, Fraccalvieri D, Bonati L, Xie Q, Zhao B. Hu Q, et al. PLoS One. 2013 Jun 11;8(6):e66258. doi: 10.1371/journal.pone.0066258. Print 2013. PLoS One. 2013. PMID: 23776647 Free PMC article. - Exactly the same but different: promiscuity and diversity in the molecular mechanisms of action of the aryl hydrocarbon (dioxin) receptor.
Denison MS, Soshilov AA, He G, DeGroot DE, Zhao B. Denison MS, et al. Toxicol Sci. 2011 Nov;124(1):1-22. doi: 10.1093/toxsci/kfr218. Epub 2011 Sep 9. Toxicol Sci. 2011. PMID: 21908767 Free PMC article. Review. - Khellin and visnagin differentially modulate AHR signaling and downstream CYP1A activity in human liver cells.
Vrzal R, Frauenstein K, Proksch P, Abel J, Dvorak Z, Haarmann-Stemmann T. Vrzal R, et al. PLoS One. 2013 Sep 19;8(9):e74917. doi: 10.1371/journal.pone.0074917. eCollection 2013. PLoS One. 2013. PMID: 24069365 Free PMC article. - Access Path to the Ligand Binding Pocket May Play a Role in Xenobiotics Selection by AhR.
Szöllősi D, Erdei Á, Gyimesi G, Magyar C, Hegedűs T. Szöllősi D, et al. PLoS One. 2016 Jan 4;11(1):e0146066. doi: 10.1371/journal.pone.0146066. eCollection 2016. PLoS One. 2016. PMID: 26727491 Free PMC article.
References
- Schmidt JV, Bradfield CA. Ah receptor signaling pathways. Annu. ReV. Cell DeV. Biol. 1996;12:55–89. - PubMed
- Denison MS, Elferink CF, Phelan D. The Ah receptor signal transduction pathway. In: Denison MS, Helferich WG, editors. Toxicant-Receptor Interactions in the Modulation of Signal Transduction and Gene Expression. Philadelphia, PA: Taylor and Francis; 1998. pp. 3–33.
- Ma Q. Induction of CYP1A1. The AhR/DRE paradigm: Transcription, receptor regulation, and expanding biological roles. Curr. Drug Metab. 2001;2:149–164. - PubMed
- Denison MS, Seidel SD, Rogers WJ, Ziccardi M, Winter GM, Heath-Pagliuso S. Natural and synthetic ligands for the Ah receptor. In: Puga A, Wallace KB, editors. Molecular Biology Approaches to Toxicology. Philadelphia, PA: Taylor & Francis; 1998. pp. 393–410.
- Denison MS, Heath-Pagliuso S. The Ah receptor: a regulator of the biochemical and toxicological actions of structurally diverse chemicals. Bull. EnViron. Contam. Toxicol. 1998;61:557–568. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- F32 ES005707/ES/NIEHS NIH HHS/United States
- R01 ES007685-10/ES/NIEHS NIH HHS/United States
- R01 ES007685/ES/NIEHS NIH HHS/United States
- P30 ES005707-159002/ES/NIEHS NIH HHS/United States
- P30 ES005707/ES/NIEHS NIH HHS/United States
- ES07685/ES/NIEHS NIH HHS/United States
- ES05707/ES/NIEHS NIH HHS/United States
LinkOut - more resources
Full Text Sources