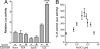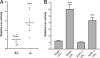The aryl hydrocarbon receptor is activated by modified low-density lipoprotein - PubMed (original) (raw)
The aryl hydrocarbon receptor is activated by modified low-density lipoprotein
Brian J McMillan et al. Proc Natl Acad Sci U S A. 2007.
Abstract
Endogenous activation of the aryl hydrocarbon receptor (AHR) is required for normal vascular development. This biology led us to investigate the interplay between the AHR and vascular physiology by using an in vitro model of fluid shear stress. Using this system, we show that fluid flow induces a robust AHR-mediated increase in CYP1 expression. Furthermore, we demonstrate that incubation with sheared bovine or human sera is sufficient for AHR activation, indicating that direct cellular exposure to shear stress is not required for this response. Fractionation of sera by size and density revealed the AHR-activating factor to be low-density lipoprotein (LDL). Purified LDL (0.1 mg/ml) from sheared sera induces a 6-fold increase in AHR-mediated signaling as compared with LDL purified from static sera. Similar results were obtained by exposing a purified fraction of LDL to fluid flow, suggesting that shear stress is capable of directly modifying LDL structure and/or function. In addition, we show that LDL can be converted to an AHR-activating species by conventional methods of lipoprotein modification, such as NaOCl oxidation. Finally, we demonstrate that an increased level of AHR-activating LDL is present in the sera of AHR null mice as compared with heterozygous littermates, suggesting a role for the Ahr locus in the physiological response to modified LDL in vivo. Overall, these data demonstrate a previously undescribed relationship between LDL modification and AHR biology and provide a potential explanation for the vascular abnormalities observed in AHR null mice.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
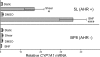
Fig. 1.
Shear stress induction of CYP1A1 depends on the AHR. The rat hepatoma 5L cell line and its derived AHR null line, BP8, were exposed in duplicate to 12 dyne/cm2 shear stress, 10 μM βNF, or control conditions for 16 h. CYP1A1 mRNA was quantitated by qRT-PCR. The relative change in mRNA expression was calculated as 2̂(ΔCT). Error bars indicate the minimum and maximum data points. This experiment was repeated, with virtually identical results. ∗, P < 0.05; ∗∗∗, P < 0.001.
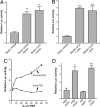
Fig. 2.
Sheared serum induces DRE-mediated transcription. (A) The H1L6.1c3 cell line, which harbors a stably integrated DRE-driven luciferase reporter, was grown to confluency on fibronectin-coated slides. In the first part of the experiment, cells (n = 3) were directly exposed to fluid shear stress (direct shear stress) or static conditions (static media) for 24 h. The growth medium “conditioned” by this experiment was then used to treat confluent slides of H1L6.1c3 cells for 24 h. (B) Media were conditioned by shear stress by using perfusion chambers that contained slides with confluent Hepa1c1c7 cells (media conditioned + cells) or slides without cells (media conditioned − cells) for 8 h. Each medium was then used to treat H1L6.1c3 cells (n = 3) for 20 h. (C) A DMEM solution supplemented with 60% vol/vol FBS (DMEM+FBS) and a nonsupplemented DMEM solution were sheared for 2 h. After exposure to shear stress, static FBS was added to the sheared DMEM solution to 60% of total volume. Dilutions of the sheared DMEM+FBS (triangles) and sheared DMEM + static FBS (circles) were used to treat H1L6.1c3 cells (n = 3) for 20 h. (D) HS and FBS were exposed to shear stress for 2 h. H1L6.1c3 cells (n = 3) were treated with a 20% vol/vol (v/v) dilution of each serum for 20 h and then assayed for luciferase activity. ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P 0.001.
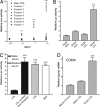
Fig. 3.
The AHR is activated by sheared LDL. (A) Sheared FBS was separated by MW using a Superose 6 gel-filtration resin. Seven total fractions were collected and used to treat H1L6.1c3 cells (n = 3) at the indicated concentrations for 18 h. (B) Static and sheared sera were fractionated by sequential density ultracentrifugation. H1L6.1c3 cells (n = 6) were treated for 20 h with 0.1 mg/ml HDL or LDL from each serum and then assayed for luciferase activity. (C) LDL was purified from static serum (black bars) or from serum exposed to shear stress for 18 h at 4C (hatched bar). An aliquot of LDL purified from the static serum was then diluted in PBS and exposed to the same shear stress conditions. Each LDL treatment group was then used to treat H1L6.1c3 cells (n = 5) for 20 h. Cells were treated with 2 μM βNF as a positive control. (D) The EOMA cell line (n = 4) was treated with 0.1 mg/ml static or sheared LDL or an equal volume of PBS for 20 h. The relative level of Cyp1a1 mRNA in each treatment group was determined by using qRT-PCR. ∗∗∗, P < 0.001.
Fig. 4.
The AHR is activated by NaOCl-modified LDL. (A) LDL, BSA, and apoB (all 0.2 mg/ml in PBS) were preincubated in the presence or absence of 100 μM NaOCl for 1 h at 37°C. After extensive dialysis of each sample, H1L6.1c3 cells (n = 4) were treated with 0.1 mg/ml of each sample in serum-free DMEM for 18 h. Statistical comparisons were performed between each NaOCl-modified substrate and its untreated control. (B) LDL was preincubated with the indicated concentration of NaOCl for 1 h at 37°C; a control sample of LDL was incubated without NaOCl. After extensive dialysis, each LDL sample (0.1 mg/ml) was used to treat H1L6.1c3 cells (n = 4) for 18 h. ∗∗∗, P < 0.001.
Fig. 5.
AHR null sera contains AHR-activating LDL. (A) Sera were isolated from nine age-matched AHR null (triangles) or heterozygous mice (squares) and used to treat H1L6.1c3 cells (n = 3) at a 30% vol/vol concentration for 20 h. The horizontal line represents the mean luciferase activity of each group. (B) LDL was purified from the sera of _Ahr_−/− and Ahr+/− littermates. H1L6.1c3 cells (n = 6) were treated with 0.1 mg/ml LDL from each mouse genotype or with LDL purified from static or sheared FBS for 20 h and then assayed for luciferase activity. ∗∗∗, P < 0.01.
Similar articles
- Modulation of aryl hydrocarbon receptor target genes in circulating lymphocytes from dairy cows bred in a dioxin-like PCB contaminated area.
Girolami F, Spalenza V, Carletti M, Sacchi P, Rasero R, Nebbia C. Girolami F, et al. Sci Total Environ. 2013 Apr 15;450-451:7-12. doi: 10.1016/j.scitotenv.2013.01.095. Epub 2013 Feb 27. Sci Total Environ. 2013. PMID: 23454571 - Benzyl butyl phthalate induces necrosis by AhR mediation of CYP1B1 expression in human granulosa cells.
Chen HS, Chiang PH, Wang YC, Kao MC, Shieh TH, Tsai CF, Tsai EM. Chen HS, et al. Reprod Toxicol. 2012 Jan;33(1):67-75. doi: 10.1016/j.reprotox.2011.11.004. Epub 2011 Nov 25. Reprod Toxicol. 2012. PMID: 22138065 - Estrogen receptor subtype- and promoter-specific modulation of aryl hydrocarbon receptor-dependent transcription.
Wihlén B, Ahmed S, Inzunza J, Matthews J. Wihlén B, et al. Mol Cancer Res. 2009 Jun;7(6):977-86. doi: 10.1158/1541-7786.MCR-08-0396. Epub 2009 May 26. Mol Cancer Res. 2009. PMID: 19470599 - Cytochrome P450 1 family and cancers.
Go RE, Hwang KA, Choi KC. Go RE, et al. J Steroid Biochem Mol Biol. 2015 Mar;147:24-30. doi: 10.1016/j.jsbmb.2014.11.003. Epub 2014 Nov 6. J Steroid Biochem Mol Biol. 2015. PMID: 25448748 Review. - Induction of cellular oxidative stress by aryl hydrocarbon receptor activation.
Dalton TP, Puga A, Shertzer HG. Dalton TP, et al. Chem Biol Interact. 2002 Sep 20;141(1-2):77-95. doi: 10.1016/s0009-2797(02)00067-4. Chem Biol Interact. 2002. PMID: 12213386 Review.
Cited by
- A Novel Aryl Hydrocarbon Receptor Antagonist HBU651 Ameliorates Peripheral and Hypothalamic Inflammation in High-Fat Diet-Induced Obese Mice.
Kang S, Lee AG, Im S, Oh SJ, Yoon HJ, Park JH, Pak YK. Kang S, et al. Int J Mol Sci. 2022 Nov 28;23(23):14871. doi: 10.3390/ijms232314871. Int J Mol Sci. 2022. PMID: 36499198 Free PMC article. - The search for endogenous activators of the aryl hydrocarbon receptor.
Nguyen LP, Bradfield CA. Nguyen LP, et al. Chem Res Toxicol. 2008 Jan;21(1):102-16. doi: 10.1021/tx7001965. Epub 2007 Dec 13. Chem Res Toxicol. 2008. PMID: 18076143 Free PMC article. Review. - Pharmacological blockage of the AHR-CYP1A1 axis: a call for in vivo evidence.
Coelho NR, Pimpão AB, Correia MJ, Rodrigues TC, Monteiro EC, Morello J, Pereira SA. Coelho NR, et al. J Mol Med (Berl). 2022 Feb;100(2):215-243. doi: 10.1007/s00109-021-02163-2. Epub 2021 Nov 20. J Mol Med (Berl). 2022. PMID: 34800164 Free PMC article. Review. - TCF21 and the environmental sensor aryl-hydrocarbon receptor cooperate to activate a pro-inflammatory gene expression program in coronary artery smooth muscle cells.
Kim JB, Pjanic M, Nguyen T, Miller CL, Iyer D, Liu B, Wang T, Sazonova O, Carcamo-Orive I, Matic LP, Maegdefessel L, Hedin U, Quertermous T. Kim JB, et al. PLoS Genet. 2017 May 8;13(5):e1006750. doi: 10.1371/journal.pgen.1006750. eCollection 2017 May. PLoS Genet. 2017. PMID: 28481916 Free PMC article. - Deficiency of Adipose Aryl Hydrocarbon Receptor Protects against Diet-Induced Metabolic Dysfunction through Sexually Dimorphic Mechanisms.
Haque N, Ojo ES, Krager SL, Tischkau SA. Haque N, et al. Cells. 2023 Jun 29;12(13):1748. doi: 10.3390/cells12131748. Cells. 2023. PMID: 37443781 Free PMC article.
References
- Gu Y-Z, Hogenesch J, Bradfield C. Annu Rev Pharmacol Toxicol. 2000;40:519–561. - PubMed
- Nebert DW, Dalton TP, Okey AB, Gonzalez FJ. J Biol Chem. 2004;279:23847–23850. - PubMed
- Schrenk D. Biochem Pharmacol. 1998;55:1155–1162. - PubMed
- Lahvis G, Pyzalski RW, Glover E, Pitot HC, McElwee MK, Bradfield CA. Mol Pharmacol. 2005;67:714–720. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 CA014520/CA/NCI NIH HHS/United States
- R37 ES005703/ES/NIEHS NIH HHS/United States
- R37-ES05703/ES/NIEHS NIH HHS/United States
- P30-CA14520/CA/NCI NIH HHS/United States
- R01 ES005703/ES/NIEHS NIH HHS/United States
- T32-CA009135/CA/NCI NIH HHS/United States
- T32 CA009135/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources