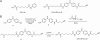Activity-based probes for proteomic profiling of histone deacetylase complexes - PubMed (original) (raw)
Activity-based probes for proteomic profiling of histone deacetylase complexes
Cleo M Salisbury et al. Proc Natl Acad Sci U S A. 2007.
Abstract
Histone deacetylases (HDACs) are key regulators of gene expression that require assembly into larger protein complexes for activity. Efforts to understand how associated proteins modulate the function of HDACs would benefit from new technologies that evaluate HDAC activity in native biological systems. Here, we describe an active site-directed chemical probe for profiling HDACs in native proteomes and live cells. This probe, designated SAHA-BPyne, contains structural elements of the general HDAC inhibitor suberoylanilide hydroxamic acid (SAHA), as well as benzophenone and alkyne moieties to effect covalent modification and enrichment of HDACs, respectively. Both class I and II HDACs were identified as specific targets of SAHA-BPyne in proteomes. Interestingly, multiple HDAC-associated proteins were also enriched by SAHA-BPyne, even after denaturation of probe-labeled proteomes. These data indicate that certain HDAC-associated proteins are directly modified by SAHA-BPyne, placing them in close proximity to HDAC active sites where they would be primed to regulate substrate recognition and activity. We further show that SAHA-BPyne can be used to measure differences in HDAC content and complex assembly in human disease models. This chemical proteomics probe should thus prove valuable for profiling both the activity state of HDACs and the binding proteins that regulate their function.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
Design and synthesis of the HDAC activity-based probe SAHA-BPyne. (A) Structures of the general HDAC inhibitor SAHA and SAHA-BPyne. (B) Synthetic scheme for preparation of SAHA-BPyne.
Fig. 2.
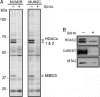
ABPP of melanoma cell proteomes with the SAHA-BPyne probe. (A) Soluble proteomes from the melanoma lines MUM2B and MUM2C were incubated with 100 nM SAHA-BPyne probe in the presence or absence of excess SAHA (10 μM) as a competitor. Probe targets were detected by UV-irradiation, followed by click chemistry with a rhodamine-azide tag, SDS/PAGE analysis, and in-gel fluorescence scanning (fluorescent gel shown in grayscale). Multiple SAHA-sensitive targets were detected (arrows). These proteins were identified as HDACs 1 and 2 (60-kDa doublet) and MBD3 (38-kDa band). (B) Confirmation that SAHA-BPyne targets both HDACs (HDAC2) and HDAC-associated (CoREST, MTA2) proteins. Shown are Western blots of proteins enriched from melanoma proteomes by treatment with SAHA-BPyne (or SAHA-BPyne plus excess SAHA), click conjugation to biotin-azide, and enrichment on avidin beads.
Fig. 3.
Alterations in HDACs and HDAC complexes between aggressive and nonaggressive melanoma lines. (A) ABPP-MudPIT with SAHA-BPyne identified lower and higher levels of HDAC6 and CoREST, respectively, in the aggressive melanoma line MUM2B compared with the less aggressive MUM2C line. P < 0.01, for levels of proteins between MUM2B and MUM2C cells. (B) Western blotting of soluble proteomes from melanoma cells confirmed that HDAC6 is more highly expressed in MUM2C cells (Upper). In contrast, equivalent levels of CoREST were observed in MUM2B and MUM2C soluble proteomes (Lower). (C) Western blotting analysis of avidin-enriched MUM2B and MUM2C proteomes treated with SAHA-BPyne revealed stronger CoREST signals in the former samples, indicating higher levels of CoREST in HDAC complexes from MUM2B cells.
Fig. 4.
Profiling HDAC complexes in living cancer cells with SAHA-BPyne. Cultured preparations of MDA-MB-231 cells were treated with 500 nM SAHA-BPyne probe in the presence or absence of excess SAHA (10 μM) and irradiated with UV light for various times. Cells were washed, scraped, and homogenized, followed by click chemistry with a rhodamine-azide tag, SDS/PAGE analysis, and in-gel fluorescence scanning (fluorescent gel shown in grayscale). Multiple SAHA-sensitive targets were detected, including those previously identified in in vitro preparations as HDAC1, HDAC2, and MBD3 (single arrowheads) and those that are more strongly labeled in living cells (double arrowheads). Labeling of the corresponding in vitro proteomic preparations of MDA-MB-231 cells is shown for reference.
Fig. 5.
Model for SAHA-BPyne labeling of HDACs and HDAC-associated proteins. Incubation of proteomes with SAHA-BPyne results in selective binding to HDACs, which exist as parts of large multiprotein complexes. The BP group of SAHA-BPyne rests on the outer rim of HDAC active sites, resulting in UV light-induced photocross-linking to both HDACs and proximally associated proteins (green). More distally associated proteins (yellow) do not react with the probe. Proteome denaturation and click chemistry with a biotin-azide tag enables identification of SAHA-BPyne-labeled proteins by ABPP-MudPIT methods (sequential avidin enrichment, on-bead trypsin digestion, and shotgun LC-MS/MS analysis).
Similar articles
- Optimization of activity-based probes for proteomic profiling of histone deacetylase complexes.
Salisbury CM, Cravatt BF. Salisbury CM, et al. J Am Chem Soc. 2008 Feb 20;130(7):2184-94. doi: 10.1021/ja074138u. Epub 2008 Jan 25. J Am Chem Soc. 2008. PMID: 18217751 - SAHA Capture Compound--a novel tool for the profiling of histone deacetylases and the identification of additional vorinostat binders.
Fischer JJ, Michaelis S, Schrey AK, Diehl A, Graebner OY, Ungewiss J, Horzowski S, Glinski M, Kroll F, Dreger M, Koester H. Fischer JJ, et al. Proteomics. 2011 Oct;11(20):4096-104. doi: 10.1002/pmic.201000717. Epub 2011 Sep 7. Proteomics. 2011. PMID: 21898820 - Design and development of histone deacetylase (HDAC) chemical probes for cell-based profiling.
Albrow VE, Grimley RL, Clulow J, Rose CR, Sun J, Warmus JS, Tate EW, Jones LH, Storer RI. Albrow VE, et al. Mol Biosyst. 2016 May 24;12(6):1781-9. doi: 10.1039/c6mb00109b. Mol Biosyst. 2016. PMID: 27021930 - Zn(II)-dependent histone deacetylase inhibitors: suberoylanilide hydroxamic acid and trichostatin A.
Codd R, Braich N, Liu J, Soe CZ, Pakchung AA. Codd R, et al. Int J Biochem Cell Biol. 2009 Apr;41(4):736-9. doi: 10.1016/j.biocel.2008.05.026. Epub 2008 Aug 3. Int J Biochem Cell Biol. 2009. PMID: 18725319 Review. - Chemical tools for probing histone deacetylase (HDAC) activity.
Minoshima M, Kikuchi K. Minoshima M, et al. Anal Sci. 2015;31(4):287-92. doi: 10.2116/analsci.31.287. Anal Sci. 2015. PMID: 25864671 Review.
Cited by
- Determining target engagement in living systems.
Simon GM, Niphakis MJ, Cravatt BF. Simon GM, et al. Nat Chem Biol. 2013 Apr;9(4):200-5. doi: 10.1038/nchembio.1211. Nat Chem Biol. 2013. PMID: 23508173 Free PMC article. - Chemical approaches to study metabolic networks.
Medina-Cleghorn D, Nomura DK. Medina-Cleghorn D, et al. Pflugers Arch. 2013 Mar;465(3):427-40. doi: 10.1007/s00424-012-1201-0. Epub 2013 Jan 8. Pflugers Arch. 2013. PMID: 23296751 Free PMC article. Review. - Roles of histone deacetylases in epigenetic regulation: emerging paradigms from studies with inhibitors.
Delcuve GP, Khan DH, Davie JR. Delcuve GP, et al. Clin Epigenetics. 2012 Mar 12;4(1):5. doi: 10.1186/1868-7083-4-5. Clin Epigenetics. 2012. PMID: 22414492 Free PMC article. - Translating HDAC inhibitors in Friedreich's ataxia.
Soragni E, Gottesfeld JM. Soragni E, et al. Expert Opin Orphan Drugs. 2016;4(9):961-970. doi: 10.1080/21678707.2016.1215910. Epub 2016 Jul 31. Expert Opin Orphan Drugs. 2016. PMID: 28392990 Free PMC article. - Development of activity-based probes for the protein deacylase Sirt1.
Goetz CJ, Sprague DJ, Smith BC. Goetz CJ, et al. Bioorg Chem. 2020 Nov;104:104232. doi: 10.1016/j.bioorg.2020.104232. Epub 2020 Aug 26. Bioorg Chem. 2020. PMID: 32911193 Free PMC article.
References
- Kuo MH, Allis CD. BioEssays. 1998;20:615–626. - PubMed
- Minucci S, Pelicci PG. Nat Rev Cancer. 2006;6:38–51. - PubMed
- Blander G, Guarente L. Annu Rev Biochem. 2004;73:417–435. - PubMed
- Marks P, Rifkind RA, Richon VM, Breslow R, Miller T, Kelly WK. Nat Rev Cancer. 2001;1:194–202. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources