Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci - PubMed (original) (raw)
Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci
Joost J B Keurentjes et al. Proc Natl Acad Sci U S A. 2007.
Abstract
Accessions of a plant species can show considerable genetic differences that are analyzed effectively by using recombinant inbred line (RIL) populations. Here we describe the results of genome-wide expression variation analysis in an RIL population of Arabidopsis thaliana. For many genes, variation in expression could be explained by expression quantitative trait loci (eQTLs). The nature and consequences of this variation are discussed based on additional genetic parameters, such as heritability and transgression and by examining the genomic position of eQTLs versus gene position, polymorphism frequency, and gene ontology. Furthermore, we developed an approach for genetic regulatory network construction by combining eQTL mapping and regulator candidate gene selection. The power of our method was shown in a case study of genes associated with flowering time, a well studied regulatory network in Arabidopsis. Results that revealed clusters of coregulated genes and their most likely regulators were in agreement with published data, and unknown relationships could be predicted.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
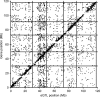
Fig. 1.
Distribution of mapped genes versus the position of their accompanying eQTL. Positions of detected eQTL are plotted against the position of the gene for which that eQTL was found. Chromosomal borders are depicted as horizontal and vertical lines. Mb, megabase.
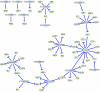
Fig. 2.
Regulatory network of genes involved in the transition to flowering. Flower genes (green dots) are connected to their most likely regulator (blue dots) by directional edges. Arrows, stimulative regulation; bars, repressive regulation.
Similar articles
- Mapping quantitative trait loci for freezing tolerance in a recombinant inbred line population of Arabidopsis thaliana accessions Tenela and C24 reveals REVEILLE1 as negative regulator of cold acclimation.
Meissner M, Orsini E, Ruschhaupt M, Melchinger AE, Hincha DK, Heyer AG. Meissner M, et al. Plant Cell Environ. 2013 Jul;36(7):1256-67. doi: 10.1111/pce.12054. Epub 2013 Jan 17. Plant Cell Environ. 2013. PMID: 23240770 - Quantitative trait loci and candidate genes underlying genotype by environment interaction in the response of Arabidopsis thaliana to drought.
El-Soda M, Kruijer W, Malosetti M, Koornneef M, Aarts MG. El-Soda M, et al. Plant Cell Environ. 2015 Mar;38(3):585-99. doi: 10.1111/pce.12418. Epub 2014 Sep 3. Plant Cell Environ. 2015. PMID: 25074022 - An expression quantitative trait loci-guided co-expression analysis for constructing regulatory network using a rice recombinant inbred line population.
Wang J, Yu H, Weng X, Xie W, Xu C, Li X, Xiao J, Zhang Q. Wang J, et al. J Exp Bot. 2014 Mar;65(4):1069-79. doi: 10.1093/jxb/ert464. Epub 2014 Jan 13. J Exp Bot. 2014. PMID: 24420573 Free PMC article. - Expression quantitative trait loci analysis in plants.
Druka A, Potokina E, Luo Z, Jiang N, Chen X, Kearsey M, Waugh R. Druka A, et al. Plant Biotechnol J. 2010 Jan;8(1):10-27. doi: 10.1111/j.1467-7652.2009.00460.x. Plant Biotechnol J. 2010. PMID: 20055957 Review. - Quantification of variation in expression networks.
Kliebenstein DJ. Kliebenstein DJ. Methods Mol Biol. 2009;553:227-45. doi: 10.1007/978-1-60327-563-7_11. Methods Mol Biol. 2009. PMID: 19588108 Review.
Cited by
- Genetical Genomics Reveals Large Scale Genotype-By-Environment Interactions in Arabidopsis thaliana.
Snoek LB, Terpstra IR, Dekter R, Van den Ackerveken G, Peeters AJ. Snoek LB, et al. Front Genet. 2013 Jan 10;3:317. doi: 10.3389/fgene.2012.00317. eCollection 2012. Front Genet. 2013. PMID: 23335938 Free PMC article. - Systems genetics in "-omics" era: current and future development.
Li H. Li H. Theory Biosci. 2013 Mar;132(1):1-16. doi: 10.1007/s12064-012-0168-x. Epub 2012 Nov 9. Theory Biosci. 2013. PMID: 23138757 Review. - The role of regulatory variation in complex traits and disease.
Albert FW, Kruglyak L. Albert FW, et al. Nat Rev Genet. 2015 Apr;16(4):197-212. doi: 10.1038/nrg3891. Epub 2015 Feb 24. Nat Rev Genet. 2015. PMID: 25707927 Review. - Transcriptome-referenced association study of clove shape traits in garlic.
Chen X, Liu X, Zhu S, Tang S, Mei S, Chen J, Li S, Liu M, Gu Y, Dai Q, Liu T. Chen X, et al. DNA Res. 2018 Dec 1;25(6):587-596. doi: 10.1093/dnares/dsy027. DNA Res. 2018. PMID: 30084885 Free PMC article. - Intra- and inter-individual genetic differences in gene expression.
Cowley MJ, Cotsapas CJ, Williams RB, Chan EK, Pulvers JN, Liu MY, Luo OJ, Nott DJ, Little PF. Cowley MJ, et al. Mamm Genome. 2009 May;20(5):281-95. doi: 10.1007/s00335-009-9181-x. Epub 2009 May 8. Mamm Genome. 2009. PMID: 19424753 Free PMC article.
References
- Jansen RC, Nap JP. Trends Genet. 2001;17:388–391. - PubMed
- Brem RB, Yvert G, Clinton R, Kruglyak L. Science. 2002;296:752–755. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources