The SRA methyl-cytosine-binding domain links DNA and histone methylation - PubMed (original) (raw)
Comparative Study
The SRA methyl-cytosine-binding domain links DNA and histone methylation
Lianna M Johnson et al. Curr Biol. 2007.
Abstract
Epigenetic gene silencing suppresses transposon activity and is critical for normal development . Two common epigenetic gene-silencing marks are DNA methylation and histone H3 lysine 9 dimethylation (H3K9me2). In Arabidopsis thaliana, H3K9me2, catalyzed by the methyltransferase KRYPTONITE (KYP/SUVH4), is required for maintenance of DNA methylation outside of the standard CG sequence context. Additionally, loss of DNA methylation in the met1 mutant correlates with a loss of H3K9me2. Here we show that KYP-dependent H3K9me2 is found at non-CG methylation sites in addition to those rich in CG methylation. Furthermore, we show that the SRA domain of KYP binds directly to methylated DNA, and SRA domains with missense mutations found in loss-of-function kyp mutants have reduced binding to methylated DNA in vitro. These data suggest that DNA methylation is required for the recruitment or activity of KYP and suggest a self-reinforcing loop between histone and DNA methylation. Lastly, we found that SRA domains from two Arabidopsis SRA-RING proteins also bind methylated DNA and that the SRA domains from KYP and SRA-RING proteins prefer methylcytosines in different sequence contexts. Hence, unlike the methyl-binding domain (MBD), which binds only methylated-CpG sequences, the SRA domain is a versatile new methyl-DNA-binding motif.
Figures
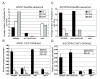
Figure 1
Correlation of DNA and histone methylation in Arabidopsis. (A) Bisulfite sequencing data for wild type, drm1 drm2 cmt3 and met1 at AtSN1, expressed as the average number of methylated CG, CNG or CNN residues per clone. (B) ChIP analysis using antibodies specific for H3K9me2 from the wild type, drm1 drm2 cmt3, met1 and kyp-6. AtSN1 was amplified from the IP and no antibody control (no AB). ACTIN was amplified from the IP. The relative quantity precipitated for each sample was derived from a standard curve of input DNA. The average of two biological replicas done in duplicate is shown with standard deviation. (C) Bisulfite sequencing data from AtCOPIA4. The remaining non-CG methylation in the drm1 drm2 cmt3 triple mutant is discussed in reference [19]. (D) ChIP analysis of H3K9me2 at AtCOPIA4. The average of two biological replicas done in duplicate is shown with standard deviation.
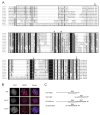
Figure 2
The SRA domain. (A) ClustalW alignment of the SRA domains from the nine SUVH genes, two Arabidopsis SRA- and RING-domain-containing proteins, mouse np95, and human ICBP90. The site of the KYP S200F[4] mutation is indicated with “#”, the E208K mutation is indicated with a “*”, and the R260H[4] mutation is indicated with a “+”. Arrows show the SRA domain identified by SMART and the boxed in amino acids are those included in the GST-SUVH6-SRA fusion protein. The association of SRA domains with SET domain proteins is unique to the plant kingdom. (B) Immunofluorescence of H3K9me2 compared to DAPI in nuclei isolated from wild type, kyp-2 (null allele), and kyp-5 (SRA mutation E208K). (C) Regions of the SUVH genes contained in the various constructs analyzed in Figure 3.
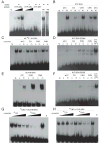
Figure 3
KYP SRA domain preferentially binds methylated DNA. (A) Mobility shift assays using increasing amounts of GST-KYP-SRA (KYP-SRA) (25, 50 and 100 nM) added to a binding reaction with either a methylated (m) or unmethylated (u) all-C double stranded oligonucleotide probe. GST-KYP-SET (KYP-SET) was added at 100 nM in the indicated lanes. Anti-GST antibodies (AB) were added to the binding reaction shown in the last lane to yield a super-shifted complex. KYP-SRA (B), KYP-SRA(E208K) (D) or KYP-SRA(S200F) (F) was bound to oligonucleotides with cytosines methylated in all sequences contexts (all-mC), or in a CG, CNG, or CNN context. In D and F the last lane is GST-KYP-SRA bound to the all-mC probe used as a positive control. (C) Competition assays were performed by addition of the indicated unlabeled DNA, added in 1000 fold excess to the all-mC probe. (E) GST-SUVH6-SRA (10 nM) was bound to the indicated probes. (G-H) GST-SUVH6-SRA (10 nM) or GST-KYP-SRA (100 nM) was bound to the methylated CNG probe with either no competitor (-), or 100x, 300x, 1000x, or 3000x unlabeled mCG or mCNG oligonucleotides. The lane indicated by fp (free probe) has no added protein.
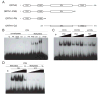
Figure 4
Binding of the ORTH proteins to methylated DNA is dependent on the SRA domain. (A) Diagram of the different regions of ORTH1 or ORTH2 fused to GST and used in mobility shift assays. (B) Mobility shift assays were done with the indicated protein using either unmethylated or methylated CG probes. *The last line in each panel contains binding of GST-PNS to methylated CG super-shifted by addition of anti-GST antibody. (C) Mobility shift assays were performed with increasing amounts (35, 70, and 140 nM) of GST-PNS binding to mCG, mCNG, or mCNN. (D). Mobility shift assays with increasing amounts of GST-PNS (70, 140, and 280 nM) or GST-KYP (77, 154, and 308 nM) with methylated (Methylated) or unmethylated (U) CG. The first lane in all panels is the free probe (fp).
Similar articles
- Mechanism of DNA methylation-directed histone methylation by KRYPTONITE.
Du J, Johnson LM, Groth M, Feng S, Hale CJ, Li S, Vashisht AA, Wohlschlegel JA, Patel DJ, Jacobsen SE. Du J, et al. Mol Cell. 2014 Aug 7;55(3):495-504. doi: 10.1016/j.molcel.2014.06.009. Epub 2014 Jul 10. Mol Cell. 2014. PMID: 25018018 Free PMC article. - Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation.
Li X, Harris CJ, Zhong Z, Chen W, Liu R, Jia B, Wang Z, Li S, Jacobsen SE, Du J. Li X, et al. Proc Natl Acad Sci U S A. 2018 Sep 11;115(37):E8793-E8802. doi: 10.1073/pnas.1809841115. Epub 2018 Aug 27. Proc Natl Acad Sci U S A. 2018. PMID: 30150382 Free PMC article. - Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase.
Ebbs ML, Bender J. Ebbs ML, et al. Plant Cell. 2006 May;18(5):1166-76. doi: 10.1105/tpc.106.041400. Epub 2006 Mar 31. Plant Cell. 2006. PMID: 16582009 Free PMC article. - DNA and histone methylation in plants.
Tariq M, Paszkowski J. Tariq M, et al. Trends Genet. 2004 Jun;20(6):244-51. doi: 10.1016/j.tig.2004.04.005. Trends Genet. 2004. PMID: 15145577 Review. - Molecular coupling of DNA methylation and histone methylation.
Hashimoto H, Vertino PM, Cheng X. Hashimoto H, et al. Epigenomics. 2010 Oct;2(5):657-69. doi: 10.2217/epi.10.44. Epigenomics. 2010. PMID: 21339843 Free PMC article. Review.
Cited by
- Mathematical model of RNA-directed DNA methylation predicts tuning of negative feedback required for stable maintenance.
Dale R, Mosher R. Dale R, et al. Open Biol. 2024 Nov;14(11):240159. doi: 10.1098/rsob.240159. Epub 2024 Nov 13. Open Biol. 2024. PMID: 39532148 Free PMC article. - Dynamic evolution of the heterochromatin sensing histone demethylase IBM1.
Zhang Y, Jang H, Luo Z, Dong Y, Xu Y, Kantamneni Y, Schmitz RJ. Zhang Y, et al. PLoS Genet. 2024 Jul 11;20(7):e1011358. doi: 10.1371/journal.pgen.1011358. eCollection 2024 Jul. PLoS Genet. 2024. PMID: 38991029 Free PMC article. - Recent Advances in Studies of Genomic DNA Methylation and Its Involvement in Regulating Drought Stress Response in Crops.
Fan Y, Sun C, Yan K, Li P, Hein I, Gilroy EM, Kear P, Bi Z, Yao P, Liu Z, Liu Y, Bai J. Fan Y, et al. Plants (Basel). 2024 May 17;13(10):1400. doi: 10.3390/plants13101400. Plants (Basel). 2024. PMID: 38794470 Free PMC article. Review. - RNA-directed DNA methylation mutants reduce histone methylation at the paramutated maize booster1 enhancer.
Hövel I, Bader R, Louwers M, Haring M, Peek K, Gent JI, Stam M. Hövel I, et al. Plant Physiol. 2024 May 31;195(2):1161-1179. doi: 10.1093/plphys/kiae072. Plant Physiol. 2024. PMID: 38366582 Free PMC article. - Allosteric crosstalk in modular proteins: Function fine-tuning and drug design.
Abhishek S, Deeksha W, Nethravathi KR, Davari MD, Rajakumara E. Abhishek S, et al. Comput Struct Biotechnol J. 2023 Oct 10;21:5003-5015. doi: 10.1016/j.csbj.2023.10.013. eCollection 2023. Comput Struct Biotechnol J. 2023. PMID: 37867971 Free PMC article. Review.
References
- Stancheva I. Caught in conspiracy: cooperation between DNA methylation and histone H3K9 methylation in the establishment and maintenance of heterochromatin. Biochem Cell Biol. 2005;83:385–395. - PubMed
- Chan SW, Henderson IR, Jacobsen SE. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat Rev Genet. 2005;6:351–360. - PubMed
- Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 GM060398/GM/NIGMS NIH HHS/United States
- R01 GM060398/GM/NIGMS NIH HHS/United States
- GM60398/GM/NIGMS NIH HHS/United States
- GM0007377-27/GM/NIGMS NIH HHS/United States
- T32 GM007377/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases