VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization - PubMed (original) (raw)
VIM1, a methylcytosine-binding protein required for centromeric heterochromatinization
Hye Ryun Woo et al. Genes Dev. 2007.
Abstract
Epigenetic regulation in eukaryotes is executed by a complex set of signaling interactions among small RNA species and chromatin marks, including histone modification and DNA methylation. We identified vim1 (VARIANT IN METHYLATION 1), an Arabidopsis mutation causing cytosine hypomethylation and decondensation of centromeres in interphase. VIM1 is a member of a small gene family, encoding proteins containing PHD, RING, and SRA (SET- and RING-associated) domains, which are found together in mammalian proteins implicated in regulation of chromatin modification, transcription, and the cell cycle. VIM1 is an unconventional methylcytosine-binding protein that interacts in vitro with 5mCpG- and 5mCpHpG-modified DNA (via its SRA domain), as well as recombinant histones (H2B, H3, H4, and HTR12) in plant extracts. VIM1 associates with methylated genomic loci in vivo and is enriched in chromocenters. Our findings suggest that VIM1 acts at the DNA methylation-histone interface to maintain centromeric heterochromatin.
Figures
Figure 1.
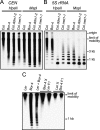
Reduced cytosine methylation of centromeric repeats in Bor-4. (A) Genomic DNA samples from the indicated genotypes were digested with the isoschizomers HpaII or MspI, and DNA blot hybridization with a 180-bp centromere repeat probe (CEN) was performed. (B) The filter shown in A was rehybridized with a 5S rRNA probe (5S rRNA). (C) A DNA blot hybridization pattern with the CEN probe after HpaII digestion demonstrates that centromeric repeat arrays hypomethylated in Bor-4 were fully remethylated in F1 hybrids resulting from reciprocal crosses between strains Bor-4 and Ler. The lane labeled Ler + Bor-4 contains a 1:1 mixture of Ler and Bor-4 HpaII-digested genomic DNA and shows the hybridization pattern expected if no remethylation occurred in the F1 hybrids.
Figure 2.
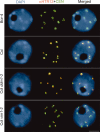
Centromeric heterochromatin is altered in Bor-4. One-hundred-eighty-base-pair centromeric repeats (CEN) were detected by FISH, and HTR12 protein was immunolocalized in interphase nuclei from root tip cells of Bor-4, Col, Col ddm1-2, and Col vim1-2 (SALK_050903) plants. The DNA was counterstained with DAPI; chromocenters are more intensely stained. Bar, 5 μm.
Figure 3.
Identification of the VIM1 gene. (A) Diagram of the intron–exon structure of the VIM1 gene and positions of T-DNA insertions in At1g57820 with allele designations. At1g57810 (reverse transcriptase pseudogene) is located between At1g57800 and At1g57820. The deletion in Bor-4 is shown as an open box (vim1-1 allele). Filled boxes represent exons and thin lines represent introns. The positions of the start and stop codon are indicated, as are the T-DNA insertion sites in At1g57820. (B) DNA blot hybridization (HpaII digest, CEN probe) analysis of T-DNA insertion mutants in At1g57820. Genomic DNA was isolated from independent plants in segregation populations. (+/+) Homozygous wild-type allele; (+/−) heterozygous for mutant allele; (−/−) homozygous for mutant allele. (C) DNA blot hybridization (HpaII digest, CEN probe) analysis demonstrating complementation of the hypomethylation phenotype of Bor-4 with a transgene expressing At1g57820 (from Col strain, coding sequence fused to a C-terminal Flag epitope tag). Analysis of six individuals in the T2 generation segregating the transgene (carrying a Basta herbicide resistance marker: [R] resistant; [S] sensitive) is shown. (D) Schematic representation of possible homologs of VIM1. The RING (Pfam PF00097), PHD (Pfam PF000628), and SRA (Pfam PF02182) domains are labeled. The mammalian proteins contain an N-terminal ubiquitin-like domain (UBL), which is absent in the VIM1-like Arabidopsis proteins, and only one RING domain. (E) Sequence alignment of SRA domains from VIM1 and three related mammalian proteins. Black shading shows identical residues and gray shading shows similar residues. Arrows denote the conserved VRV(I/V)RG and YDG motifs in SRA domains.
Figure 4.
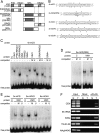
VIM1 is a methylcytosine-binding protein. (A) Schematic of the VIM1 protein and derivatives tested for methylcytosine-binding activity; amino acid coordinates are shown in the parentheses. Domain designations include PHD domain, SRA domain, and RING domains. (B) Oligonucleotide probes used in the methylcytosine-binding assays shown in C_–_E. Asterisks indicate the positions of 5mC residues. The underlined 5′-CCGG-3′ site corresponds to the HpaII assayed in Figure 1. (C) VIM1 protein derivatives were expressed in wheat germ transcription/translation extracts (C, vector-only control) and incubated with the digoxigenin-endlabeled 4x-mCG probe in the presence of 62-fold excess unlabeled competitor oligonucleotides. (U) Unmethylated; (M) methylated; (−) no competitor. EMSA of incubation products is shown; the position of the unbound free probe is indicated at the bottom of the image. The same abbreviations and symbols are used in C_–_E. (D) EMSA demonstrating the ability of full-length VIM1 to bind a methylated oligonucleotide probe based on the 180-bp centromere repeat sequence. (E) EMSA of interaction of full-length VIM1 with identical probes methylated in different cytosine contexts. (F) ChIP was performed on nuclei prepared from Col (N, nontransgenic) and transgenic (T) plants expressing Flag-VIM1. After immunoprecipitation, samples were used as templates for amplification of different genomic sequences, which are indicated at the left of the images of the resulting ethidium-stained gels. (Input) 10% of input; (Mock) precipitation in the absence of antibody; (αFlag) precipitation with anti-Flag antibody.
Figure 5.
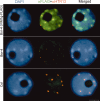
Subnuclear localization of VIM1. Immunolocalization of the epitope-tagged VIM1 (αFlag) and HTR12 (αHTR12) was performed on nuclei derived from nontransgenic Bor-4 and Col plants, as well as transgenic Bor-4 plants expressing VIM1-Flag under control of the native VIM1 promoter (VIM1g-Flag). DAPI was used as a DNA counterstain. Bar, 5 μm.
Figure 6.
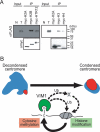
VIM1 at the DNA methylation–chromatin interface. (A) VIM1 interacts with histones. (Top panel) Immobilized myc-tagged histones were incubated with protein extracts from transgenic plants expressing Flag-VIM1, and the associated proteins were collected and detected by Western blot analysis using an anti-Flag antibody to detect Flag-VIM1. Input lanes correspond to total protein from either nontransgenic (N) or transgenic (T) plants (1/240×). The bottom panel shows detection of myc-tagged histones in the samples using an anti-myc antibody. (B) A model for the role of VIM1 in the maintenance of centromeric heterochromatin. The centromere decondensation phenotype is illustrated on the top left. (Blue) 180-bp centromere repeat; (red) HTR12. The VIM1 biochemical activities illustrated at the bottom of the figure are necessary for centromere condensation (top right). Demonstrated VIM1 activities include direct binding of methylcytosine (m–C) and histone interaction (direct or indirect). VIM1 is proposed to act as a ubiquitin (Ub) ligase to modify centromere chromatin structure through direct modification of nucleosomal histones or through modification of one or more chromatin effector proteins (X). An alteration in centromeric chromatin structure leads to DNA hypomethylation and decondensation of the centromere repeats.
Similar articles
- Plant SET- and RING-associated domain proteins in heterochromatinization.
Liu S, Yu Y, Ruan Y, Meyer D, Wolff M, Xu L, Wang N, Steinmetz A, Shen WH. Liu S, et al. Plant J. 2007 Dec;52(5):914-26. doi: 10.1111/j.1365-313X.2007.03286.x. Epub 2007 Sep 22. Plant J. 2007. PMID: 17892444 - Three SRA-domain methylcytosine-binding proteins cooperate to maintain global CpG methylation and epigenetic silencing in Arabidopsis.
Woo HR, Dittmer TA, Richards EJ. Woo HR, et al. PLoS Genet. 2008 Aug 15;4(8):e1000156. doi: 10.1371/journal.pgen.1000156. PLoS Genet. 2008. PMID: 18704160 Free PMC article. - Arabidopsis VIM proteins regulate epigenetic silencing by modulating DNA methylation and histone modification in cooperation with MET1.
Kim J, Kim JH, Richards EJ, Chung KM, Woo HR. Kim J, et al. Mol Plant. 2014 Sep;7(9):1470-1485. doi: 10.1093/mp/ssu079. Epub 2014 Jul 9. Mol Plant. 2014. PMID: 25009302 Free PMC article. - The value-added genome: building and maintaining genomic cytosine methylation landscapes.
Rangwala SH, Richards EJ. Rangwala SH, et al. Curr Opin Genet Dev. 2004 Dec;14(6):686-91. doi: 10.1016/j.gde.2004.09.009. Curr Opin Genet Dev. 2004. PMID: 15531165 Review. - [Mechanisms to control DNA methylation in Arabidopsis thaliana].
Sasaki T, Saze H, Kakutani T. Sasaki T, et al. Tanpakushitsu Kakusan Koso. 2008 Jun;53(7):809-14. Tanpakushitsu Kakusan Koso. 2008. PMID: 18536342 Review. Japanese. No abstract available.
Cited by
- Bridging the gap: unravelling plant centromeres in the telomere-to-telomere era.
Naish M. Naish M. New Phytol. 2024 Dec;244(6):2143-2149. doi: 10.1111/nph.20149. Epub 2024 Sep 27. New Phytol. 2024. PMID: 39329317 Free PMC article. Review. - Epigenetic gene regulation in plants and its potential applications in crop improvement.
Zhang H, Zhu JK. Zhang H, et al. Nat Rev Mol Cell Biol. 2025 Jan;26(1):51-67. doi: 10.1038/s41580-024-00769-1. Epub 2024 Aug 27. Nat Rev Mol Cell Biol. 2025. PMID: 39192154 Review. - The structure, function, and evolution of plant centromeres.
Naish M, Henderson IR. Naish M, et al. Genome Res. 2024 Mar 20;34(2):161-178. doi: 10.1101/gr.278409.123. Genome Res. 2024. PMID: 38485193 Free PMC article. Review. - Natural polymorphisms in ZMET2 encoding a DNA methyltransferase modulate the number of husk layers in maize.
Wang Z, Xia A, Wang Q, Cui Z, Lu M, Ye Y, Wang Y, He Y. Wang Z, et al. Plant Physiol. 2024 Jun 28;195(3):2129-2142. doi: 10.1093/plphys/kiae113. Plant Physiol. 2024. PMID: 38431291 Free PMC article. - Chromatin remodeling of histone H3 variants by DDM1 underlies epigenetic inheritance of DNA methylation.
Lee SC, Adams DW, Ipsaro JJ, Cahn J, Lynn J, Kim HS, Berube B, Major V, Calarco JP, LeBlanc C, Bhattacharjee S, Ramu U, Grimanelli D, Jacob Y, Voigt P, Joshua-Tor L, Martienssen RA. Lee SC, et al. Cell. 2023 Sep 14;186(19):4100-4116.e15. doi: 10.1016/j.cell.2023.08.001. Epub 2023 Aug 28. Cell. 2023. PMID: 37643610 Free PMC article.
References
- Alonso J.M., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Stepanova A.N., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Leisse T.J., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Kim C.J., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Chen H., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Shinn P., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Stevenson D.K., Zimmerman J., Barajas P., Cheuk R., Zimmerman J., Barajas P., Cheuk R., Barajas P., Cheuk R., Cheuk R., et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. - PubMed
- Amir R.E., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y., Van den Veyver I.B., Wan M., Tran C.Q., Francke U., Zoghbi H.Y., Wan M., Tran C.Q., Francke U., Zoghbi H.Y., Tran C.Q., Francke U., Zoghbi H.Y., Francke U., Zoghbi H.Y., Zoghbi H.Y. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999;23:185–188. - PubMed
- Arima Y., Hirota T., Bronner C., Mousli M., Fujiwara T., Niwa S., Ishikawa H., Saya H., Hirota T., Bronner C., Mousli M., Fujiwara T., Niwa S., Ishikawa H., Saya H., Bronner C., Mousli M., Fujiwara T., Niwa S., Ishikawa H., Saya H., Mousli M., Fujiwara T., Niwa S., Ishikawa H., Saya H., Fujiwara T., Niwa S., Ishikawa H., Saya H., Niwa S., Ishikawa H., Saya H., Ishikawa H., Saya H., Saya H. Down-regulation of nuclear protein ICBP90 by p53/p21Cip1/WAF1-dependent DNA-damage checkpoint signals contributes to cell cycle arrest at G1/S transition. Genes Cells. 2004;9:131–142. - PubMed
- Bechtold N., Pelletier G., Pelletier G. In planta Agrobacterium-mediated transformation of adult Arabidopsis thaliana plants by vacuum infiltration. Methods Mol. Biol. 1998;82:259–266. - PubMed
- Bonapace I.M., Latella L., Papait R., Nicassio F., Sacco A., Muto M., Crescenzi M., Di Fiore P.P., Latella L., Papait R., Nicassio F., Sacco A., Muto M., Crescenzi M., Di Fiore P.P., Papait R., Nicassio F., Sacco A., Muto M., Crescenzi M., Di Fiore P.P., Nicassio F., Sacco A., Muto M., Crescenzi M., Di Fiore P.P., Sacco A., Muto M., Crescenzi M., Di Fiore P.P., Muto M., Crescenzi M., Di Fiore P.P., Crescenzi M., Di Fiore P.P., Di Fiore P.P. Np95 is regulated by E1A during mitotic reactivation of terminally differentiated cells and is essential for S phase entry. J. Cell Biol. 2002;157:909–914. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM060380/GM/NIGMS NIH HHS/United States
- R01 GM077590/GM/NIGMS NIH HHS/United States
- R01GM077590/GM/NIGMS NIH HHS/United States
- R01GM60380/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases