Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression - PubMed (original) (raw)
. 2007 Mar;27(6):2240-52.
doi: 10.1128/MCB.02005-06. Epub 2007 Jan 22.
Janell Schelter, Julja Burchard, Miho Kibukawa, Melissa M Martin, Steven R Bartz, Jason M Johnson, Jordan M Cummins, Christopher K Raymond, Hongyue Dai, Nelson Chau, Michele Cleary, Aimee L Jackson, Michael Carleton, Lee Lim
Affiliations
- PMID: 17242205
- PMCID: PMC1820501
- DOI: 10.1128/MCB.02005-06
Transcripts targeted by the microRNA-16 family cooperatively regulate cell cycle progression
Peter S Linsley et al. Mol Cell Biol. 2007 Mar.
Abstract
microRNAs (miRNAs) are abundant, approximately 21-nucleotide, noncoding regulatory RNAs. Each miRNA may regulate hundreds of mRNA targets, but the identities of these targets and the processes they regulate are poorly understood. Here we have explored the use of microarray profiling and functional screening to identify targets and biological processes triggered by the transfection of human cells with miRNAs. We demonstrate that a family of miRNAs sharing sequence identity with miRNA-16 (miR-16) negatively regulates cellular growth and cell cycle progression. miR-16-down-regulated transcripts were enriched with genes whose silencing by small interfering RNAs causes an accumulation of cells in G(0)/G(1). Simultaneous silencing of these genes was more effective at blocking cell cycle progression than disruption of the individual genes. Thus, miR-16 coordinately regulates targets that may act in concert to control cell cycle progression.
Figures
FIG. 1.
Microarray profiling identifies a family of miRNAs that regulates cell cycle transcripts. (A) Strategy for identifying pathways regulated by miRNAs. HCT116 or DLD-1 Dicerex5 cells were transfected with miRNAs, and RNA was isolated 24 h posttransfection. Gene expression profiles were determined by competitive hybridization of amplified mRNAs from miRNA-treated versus mock-treated cells. These profiles were then examined for enrichment with transcripts containing 3′UTR seed hexamer matches or those corresponding to biological annotation categories. (B) miRNA family whose expression signature scores highly for biological annotation. Expression signatures generated by 24 miRNAs were tested for enrichment with sequences annotated with GO biological process terms and 3′UTR seed hexamer matches. Shown are the three miRNAs whose expression signatures showed significant enrichment with GO biological process sequences [E < 1E(−2)]. The highest-scoring GO biological process term, the percentage of transcripts annotated with that term (percentage of targets), and the _E_ value for enrichment are shown. For 3′UTR hexamer annotation, the percentage of transcripts containing the top-ranked hexamer match and the _E_ value for enrichment are shown. There were 18,124 total annotated sequences on the microarray, of which ∼20% contained the top-ranked 3′UTR hexamer for miR-103, miR-15a, and miR-16. The miR-103, miR-15a, and miR-16 signatures comprised 215, 549, and 557 transcripts, respectively (Table S2 in the supplemental material). (C) Kinetic separation of miR-15, −16 target regulation, and the cell cycle gene regulation phenotype. HCT116 Dicerex5 cells were transfected with luciferase siRNA (luc) or miR-106b, miR-15a, or miR-16 duplexes. RNA samples were isolated 6, 10, 14, and 24 h after transfection and were compared to RNA from mock-transfected cells. Shown is a heat-map representation of regulated genes (columns) in different experiments (rows). Samples are arranged by increasing time after transfection, from top to bottom. The color bar represents log10 expression ratios (samples from treated cells/samples from mock-treated cells) of −0.6 (teal) to +0.6 (magenta). Shown are results for 1,394 transcripts regulated with _P_ of <0.05 and a log10 expression ratio of <0 in any one experiment. The 6-h signature transcripts were required additionally to be present at a majority of later time points. Individual branches of the dendrogram were tested for enrichment with transcripts annotated with the GO biological process term mitotic cell cycle and the miR-15a-miR-16 seed region hexamer beginning at position 2. The _E_ values for enrichment with cell cycle-related transcripts were 5.5E(−26) for the branch showing enrichment with mitotic cell cycle sequences and >1E(−2) for the branch showing enrichment with miR-16 hexamers. The E values for enrichment with miR-16 hexamers were 1.1E(−80) for the branch showing enrichment with miR-16 hexamers and >1E(−2) for the branch showing enrichment with mitotic cell cycle sequences.
FIG. 2.
miR-15 and miR-16 cause the accumulation of cells at the Golgi stage of the cell cycle. (A) miR-16 triggers the accumulation of cells at a stage of the cell cycle. HCT116 Dicerex5 cells were transfected with miR-106b or miR-16. −Nocodazole, cells were analyzed for cell cycle distribution 24 h posttransfection; +Nocodazole, cells were treated with nocodazole beginning at 20 h posttransfection and analyzed for cell cycle distribution 18 h later (46 h posttransfection). 2N, cells having diploid DNA content; 4N, cells having tetraploid DNA content. miR-106b gave a cell cycle profile indistinguishable from that of mock- or luciferase-transfected cells. (B) miR-16 causes the accumulation of cells at a stage of the cell cycle rather than cell cycle arrest. HCT116 Dicerex5 cells were transfected with miR-106b (blue) or miR-16 (magenta). Cells were treated with nocodazole 24 h after transfection and analyzed at the indicated times after nocodazole addition for cell cycle distribution. Shown are the fractions of total cells in G0/G1 (2N DNA; circles) and G2/M (4N DNA; triangles) with increasing time in nocodazole. (C) The miR-16 cell cycle phenotype is seed region dependent. HCT116 Dicerex5 cells were transfected with miR-106b, miR-16, or miR-16 containing mismatches at positions 2 and 3 (2,3 mm) or 18 and 19 (18,19 mm). Cells were treated with nocodazole and analyzed as described in the legend to panel A.
FIG. 3.
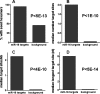
Natural forms and levels of miR-16 regulate cell cycle progression. (A) miR-16 expressed from hairpin precursors can trigger the phenotype of accumulation of cells at a stage of the cell cycle. HCT116 Dicerex5 cells were transfected with increasing concentrations of the miR-16 duplex (0.5, 1, 10, and 100 nM) or plasmids carrying miR-16 expressed as an shRNA (mR-16 hairpin) or from its endogenous locus on chromosome 13 (miR-16 locus). For transfections with plasmids, the DNA concentration was chosen to maximize transfection efficiency while minimizing toxicity (1.5 μg of DNA/2 × 105 to 3 × 105 cells in a 6-well dish). Cells were treated with nocodazole and analyzed as described in the legend to Fig. 2A. The percentages of cells in G0/G1 in different experiments were normalized so that background control and miR-16-transfected cells gave 0% and 100% of cells in G0/G1, respectively [(the percentage of G1 cells in the sample − the percentage of background G1 cells)/(the percentage of G1 miR-16-transfected cells − the percentage of background G1 cells) × 100]. For duplex transfections, we used mock-transfected cells to determine the percentage of background G1 cells. For plasmid transfections, we used cells transfected with an empty vector to determine the percentage of background G1 cells. miR-16 copy numbers (copies/20 pg of RNA) were determined by a quantitative primer extension PCR assay. miR-16 copy numbers shown are the means of quadruplicate determinations that typically differed from the mean by <15%. The results shown are representative of results from at least two experiments for each form of miR-16. (B) miR-16 and miR-106b targets are up-regulated by specific anti-miRs. HeLa cells were transfected with luciferase siRNA, miR-16, miR-106b, anti-miR-16, or anti-miR-106b. Microarray analysis was performed as described in the legend to Fig. 1C, and gene expression changes corresponding to consensus miR-16 (left panel)- and miR-106b (right panel)-down-regulated transcripts were examined. Shown is a heat-map depiction of gene expression changes in cells transfected with different miRNAs or anti-miRs. (C) Summary of miR-16 and miR-106b target regulation by specific anti-miRs in different cell lines. HeLa, TOV21G, HCT116 wild-type, and HCT116 Dicerex5 cells were transfected with anti-miR-16 (top panel) or anti-miR-106b (bottom panel). Microarray analysis was performed as described in the legend to Fig. 1C, and gene expression changes corresponding to miR-16 consensus and miR-106b consensus targets (see Table S4 in the supplemental material) were examined. For controls, we compared patterns of regulation of randomized sets of genes in luciferase siRNA-treated cells. Median change, median percentage of increase in regulation. Control sets gave 0.01% ± 0.6% change. # up-regulated, percentage of miR-16 or miR-106b consensus targets having a level of regulation of >0. Control sets gave 50% ± 5% of targets with a level of regulation of >0. Up-regulation P value, Wilcoxon signed-rank P values for the up-regulation of the indicated target sets. Mitotic cell cycle genes, transcripts down-regulated by the miR-16 duplex at 24 h and annotated with the GO biological process term mitotic cell cycle (Fig. 1C).
FIG. 4.
miR-16-down-regulated transcripts contain multiple miR-16 target sites. Properties of consensus miR-16-down-regulated transcripts (see Table S4 in the supplemental material) were compared with those of an expression level-matched background set. The significance of the differences between the groups is indicated (Wilcoxon rank-sum P values). (A) Nearly all miR-16-down-regulated transcripts contain miR-16 target sites in their 3′UTRs. Shown are the percentages of miR-16-down-regulated transcripts and background transcripts that contain sites matching miR-16 seed region hexamers in their 3′UTRs (miR-16 target sites). (B) miR-16-down-regulated transcripts contain multiple copies of miR-16 target sites in their 3′UTRs. Shown are the median numbers of miR-16 target sites in 3′UTRs of miR-16-down-regulated transcripts and background transcripts. (C) miR-16-down-regulated transcripts contain a higher density of miR-16 target sites in their 3′UTRs. Shown are the median densities of miR-16 target sites in the 3′UTRs of miR-16-down-regulated transcripts and background transcripts. (D) miR-16-down-regulated transcripts contain longer miR-16 target sites in their 3′UTRs. The longest target site per transcript was identified, and the median lengths of the longest target sites in the 3′UTRs of miR-16-down-regulated transcripts and background transcripts are shown.
FIG. 5.
miR-16-down-regulated transcripts cooperatively regulate cell cycle progression. (A) miR-16-down regulated transcripts are enriched with targets whose disruption causes the accumulation of cells in G0/G1. HCT116 Dicerex5 cells were individually transfected with siRNA pools targeting 102 transcripts containing matches to the miR-16 seed region (miR-16 targets) and 51 transcripts that did not contain miR-16 seed region matches (non-miR-16 targets). Cell cycle phenotypes were determined as described in the legend to Fig. 2. The percentages of cells in G0/G1 in different experiments were normalized as described in the legend to Fig. 3A. Before normalization, the background control had an average of 6.4% + 1.1% (mean + standard deviation) of cells in G0/G1, and among miR-16-transfected cells, 41% + 5% were in G0/G1 (results are from seven independent experiments). Plotted are normalized percentages of cells in G0/G1 for each siRNA pool (dots). The dotted line indicates a cutoff of 20%, chosen statistically to maximize the recovery of miR-16 targets while maintaining the significance of the difference between miR-16 targets and non-miR-16 targets. Non-miR-16 targets tested are listed in Table S5 in the supplemental material. (B) Cells transfected with selected siRNA pools are phenocopies of miR-16-transfected cells. HCT116 Dicerex5 cells were transfected with individual miR-16, miR-106b (control), and siRNA pools corresponding to selected miR-16-down-regulated targets (three siRNAs, each at 33 nM; 100 nM total concentration). (C) Cooperative cell cycle regulation by miR-16-down-regulated targets. Cells were transfected with individual siRNAs at 0.25 nM corresponding to selected miR-16-down-regulated targets that induce the accumulation of cells in G0/G1. Cells were also transfected with these siRNAs together as a pool (pool 3, 0.25 nM [each] siRNAs; total concentration, 1 nM). Cells were transfected with miR-16 (1 nM and 100 nM) and miR-106b (100 nM) as controls. Cell cycle phenotypes were determined as described in the legend to Fig. 2.
FIG. 6.
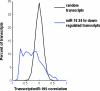
Levels of miR-16-down-regulated transcripts negatively correlate with miR-195 levels in human tumors. RNA was isolated from a series of 29 tumors and 28 adjacent uninvolved normal tissues. mRNA expression was measured using microarrays, and miR-195 levels were determined using a quantitative primer extension PCR assay. mRNA and miRNA expression levels in tumors and adjacent normal tissues were expressed as ratios of these levels to expression levels in a pool of normal samples from each tissue type. Correlations between expression level ratios for miR-195 and transcripts down-regulated 24 h after the transfection of tissue culture cells with miR-16 were calculated. As a control, correlations were also calculated for ∼200 random permutations of expression ratios (random transcripts).
Similar articles
- Coordinated regulation of cell cycle transcripts by p53-Inducible microRNAs, miR-192 and miR-215.
Georges SA, Biery MC, Kim SY, Schelter JM, Guo J, Chang AN, Jackson AL, Carleton MO, Linsley PS, Cleary MA, Chau BN. Georges SA, et al. Cancer Res. 2008 Dec 15;68(24):10105-12. doi: 10.1158/0008-5472.CAN-08-1846. Cancer Res. 2008. PMID: 19074876 - MicroRNA-34b and MicroRNA-34c are targets of p53 and cooperate in control of cell proliferation and adhesion-independent growth.
Corney DC, Flesken-Nikitin A, Godwin AK, Wang W, Nikitin AY. Corney DC, et al. Cancer Res. 2007 Sep 15;67(18):8433-8. doi: 10.1158/0008-5472.CAN-07-1585. Epub 2007 Sep 6. Cancer Res. 2007. PMID: 17823410 - Target identification of microRNAs expressed highly in human embryonic stem cells.
Li SS, Yu SL, Kao LP, Tsai ZY, Singh S, Chen BZ, Ho BC, Liu YH, Yang PC. Li SS, et al. J Cell Biochem. 2009 Apr 15;106(6):1020-30. doi: 10.1002/jcb.22084. J Cell Biochem. 2009. PMID: 19229866 - [Cellular function of microRNA-15 family].
Liu LF, Wang Y. Liu LF, et al. Sheng Li Xue Bao. 2012 Feb 25;64(1):101-6. Sheng Li Xue Bao. 2012. PMID: 22348968 Review. Chinese. - MicroRNAs and cell cycle regulation.
Carleton M, Cleary MA, Linsley PS. Carleton M, et al. Cell Cycle. 2007 Sep 1;6(17):2127-32. doi: 10.4161/cc.6.17.4641. Epub 2007 Jun 26. Cell Cycle. 2007. PMID: 17786041 Review.
Cited by
- TarBase 6.0: capturing the exponential growth of miRNA targets with experimental support.
Vergoulis T, Vlachos IS, Alexiou P, Georgakilas G, Maragkakis M, Reczko M, Gerangelos S, Koziris N, Dalamagas T, Hatzigeorgiou AG. Vergoulis T, et al. Nucleic Acids Res. 2012 Jan;40(Database issue):D222-9. doi: 10.1093/nar/gkr1161. Epub 2011 Dec 1. Nucleic Acids Res. 2012. PMID: 22135297 Free PMC article. - RNA sequence analysis defines Dicer's role in mouse embryonic stem cells.
Calabrese JM, Seila AC, Yeo GW, Sharp PA. Calabrese JM, et al. Proc Natl Acad Sci U S A. 2007 Nov 13;104(46):18097-102. doi: 10.1073/pnas.0709193104. Epub 2007 Nov 7. Proc Natl Acad Sci U S A. 2007. PMID: 17989215 Free PMC article. - Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA.
Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO. Hendrickson DG, et al. PLoS Biol. 2009 Nov;7(11):e1000238. doi: 10.1371/journal.pbio.1000238. Epub 2009 Nov 10. PLoS Biol. 2009. PMID: 19901979 Free PMC article. - The Role and Therapeutic Potential of miRNAs in Colorectal Liver Metastasis.
Sahu SS, Dey S, Nabinger SC, Jiang G, Bates A, Tanaka H, Liu Y, Kota J. Sahu SS, et al. Sci Rep. 2019 Nov 1;9(1):15803. doi: 10.1038/s41598-019-52225-2. Sci Rep. 2019. PMID: 31676795 Free PMC article. - Prediction methods for microRNA targets in bilaterian animals: Toward a better understanding by biologists.
Quillet A, Anouar Y, Lecroq T, Dubessy C. Quillet A, et al. Comput Struct Biotechnol J. 2021 Oct 18;19:5811-5825. doi: 10.1016/j.csbj.2021.10.025. eCollection 2021. Comput Struct Biotechnol J. 2021. PMID: 34765096 Free PMC article. Review.
References
- Ambros, V. 2004. The functions of animal microRNAs. Nature 431:350-355. - PubMed
- Bagga, S., J. Bracht, S. Hunter, K. Massirer, J. Holtz, R. Eachus, and A. E. Pasquinelli. 2005. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell 122:553-563. - PubMed
- Bartel, D. P., and C. Z. Chen. 2004. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat. Rev. Genet. 5:396-400. - PubMed
- Bentwich, I. 2005. Prediction and validation of microRNAs and their targets. FEBS Lett. 579:5904-5910. - PubMed
- Bentwich, I., A. Avniel, Y. Karov, R. Aharonov, S. Gilad, O. Barad, A. Barzilai, P. Einat, U. Einav, E. Meiri, E. Sharon, Y. Spector, and Z. Bentwich. 2005. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 37:766-770. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources