Large-scale phosphorylation analysis of mouse liver - PubMed (original) (raw)
Large-scale phosphorylation analysis of mouse liver
Judit Villén et al. Proc Natl Acad Sci U S A. 2007.
Abstract
Protein phosphorylation is a complex network of signaling and regulatory events that affects virtually every cellular process. Our understanding of the nature of this network as a whole remains limited, largely because of an array of technical challenges in the isolation and high-throughput sequencing of phosphorylated species. In the present work, we demonstrate that a combination of tandem phosphopeptide enrichment methods, high performance MS, and optimized database search/data filtering strategies is a powerful tool for surveying the phosphoproteome. Using our integrated analytical platform, we report the identification of 5,635 nonredundant phosphorylation sites from 2,328 proteins from mouse liver. From this list of sites, we extracted both novel and known motifs for specific Ser/Thr kinases including a "dipolar" motif. We also found that C-terminal phosphorylation was more frequent than at any other location and that the distribution of potential kinases for these sites was unique. Finally, we identified double phosphorylation motifs that may be involved in ordered phosphorylation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
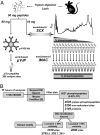
Strategy used for the large-scale identification and characterization of phosphorylation sites from mouse liver. (A) Sample preparation. Tissue homogenization and lysis was followed by trypsin digestion. Tryptic peptides (10 mg) were subjected to a two-step phosphopeptide enrichment. SCX chromatography provided a substantial enrichment in early eluting fractions. Subsequent IMAC of each fraction provided additional selective capture. In addition, 80 mg of tryptic peptides were enriched for pTyr-containing peptides by immunoaffinity purification. (B) Data processing for the SCX/IMAC experiment. MS/MS spectra from 15 analyses were searched with Sequest against the mouse protein database (DB) containing both forward (target) and reversed (decoy) sequences for FP calculations. Importantly, the target/decoy database search strategy also provided a means to establish appropriate orthogonal filtering criteria (mass deviation, enzyme specificity, solution charge state at pH 2.65, and Sequest scoring). Only two reversed-sequence peptides were found in the final filtered list of 8,529 phosphopeptides (0.02% FP rate). In total, 5,250 nonredundant sites were identified. The Ascore algorithm (26) was used to determine a probability of correct localization for each individual site. Finally, phosphorylation motifs were extracted from the data set with the Motif-X algorithm (34).
Fig. 2.
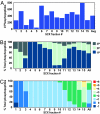
Distribution of phosphopeptides and their properties across 15 SCX fractions. (A) Phosphopeptide distribution. Shown are data for phosphopeptides identified per fraction. (B) Phosphorylation sites per peptide. Shown are percentages of phosphopeptides in each fraction containing one, two, or three phosphorylation sites. (C) Net solution charge state (pH 2.65). Shown are percentages of phosphopeptides in each fraction with calculated solution charge states between −1 and +6. SCX chromatography separates primarily based on net solution charge state, and each phosphate group subtracts one net charge from a peptide.
Fig. 3.
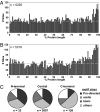
Positional distribution of phosphorylation sites. (A) Frequency of site detection with respect to protein sequence position for this study. Protein sequences were divided into 1% bins and plotted by frequency. The dashed line shows the median value. A strong trend for C-terminal (98–100% of the protein length) phosphorylation was observed. (B) This same trend was observed for the distribution of sites from the Phospho.ELM database, a curated resource containing sites from the literature. (C) Classification of phosphorylation sites into the three most general motif classes based on their position within the protein. All localized sites were classified into one of three general kinase motifs or as “other” (see Methods), and their distributions at protein ends were determined. N-terminal, within first 10 aa; C-terminal, within last 10 aa; central, within all remaining residues. C-terminal sites had different distributions than N-terminal or central sites.
Fig. 4.
Classification of localized sites (P < 0.01) into the most general kinase recognition sequence categories: acidic, basic, Pro-directed, and “others.” (A) Comparison of sequence category distributions for the sites detected in this study with the control set of all Ser and Thr residues from the same set of proteins. (B) Diverse phosphorylation patterns were observed for proteins belonging to different GO annotation cellular localization categories. (C) Examples of general phosphorylation kinase classification patterns for three functional GO categories.
Fig. 5.
Phosphorylation-specific motifs using the Motif-X algorithm (34). The data set included all sites with Ascore values of ≥19 (n = 3,439). The complete set of motifs is shown in
SI Table 4
. (A_–_E) Sequence logos for some examples of single-phosphorylation motifs where the phosphorylated residue (S or T) is centered. (A) Pro-directed motifs. (B) Basic motifs representative of CaM kinase and Akt kinase substrates, respectively. (C) Acidic motif with 56 occurrences in our data set of 630 in the entire mouse database. (D) Pro-directed motif centered on Thr with a strong preference for additional Pro residues C-terminal to the phosphate. (E) pTyr motif. (F_–_I) Examples of double-phosphorylation motifs. Secondary phosphorylated sites away from the central residue position also are phosphorylated. (F) Pro-directed with additional pSer at +4. (G) Basic upstream and acidic downstream. (H) Acidic motifs with two pSer residues. This category was the most highly represented for doubly phosphorylated peptides. (I) pThr-directed motif.
Similar articles
- Phosphoproteomic analysis of rat liver by high capacity IMAC and LC-MS/MS.
Moser K, White FM. Moser K, et al. J Proteome Res. 2006 Jan;5(1):98-104. doi: 10.1021/pr0503073. J Proteome Res. 2006. PMID: 16396499 - Occurrence and detection of phosphopeptide isomers in large-scale phosphoproteomics experiments.
Courcelles M, Bridon G, Lemieux S, Thibault P. Courcelles M, et al. J Proteome Res. 2012 Jul 6;11(7):3753-65. doi: 10.1021/pr300229m. Epub 2012 Jun 22. J Proteome Res. 2012. PMID: 22668510 - Integrative network analysis of the signaling cascades in seedling leaves of bread wheat by large-scale phosphoproteomic profiling.
Lv DW, Ge P, Zhang M, Cheng ZW, Li XH, Yan YM. Lv DW, et al. J Proteome Res. 2014 May 2;13(5):2381-95. doi: 10.1021/pr401184v. Epub 2014 Mar 28. J Proteome Res. 2014. PMID: 24679076 - Catch me if you can: mass spectrometry-based phosphoproteomics and quantification strategies.
Eyrich B, Sickmann A, Zahedi RP. Eyrich B, et al. Proteomics. 2011 Feb;11(4):554-70. doi: 10.1002/pmic.201000489. Epub 2011 Jan 11. Proteomics. 2011. PMID: 21226000 Review. - Phosphoproteomics by mass spectrometry and classical protein chemistry approaches.
Salih E. Salih E. Mass Spectrom Rev. 2005 Nov-Dec;24(6):828-46. doi: 10.1002/mas.20042. Mass Spectrom Rev. 2005. PMID: 15538747 Review.
Cited by
- Splice-specific glycine receptor binding, folding, and phosphorylation of the scaffolding protein gephyrin.
Herweg J, Schwarz G. Herweg J, et al. J Biol Chem. 2012 Apr 13;287(16):12645-56. doi: 10.1074/jbc.M112.341826. Epub 2012 Feb 17. J Biol Chem. 2012. PMID: 22351777 Free PMC article. - Nuts and bolts of the salt-inducible kinases (SIKs).
Darling NJ, Cohen P. Darling NJ, et al. Biochem J. 2021 Apr 16;478(7):1377-1397. doi: 10.1042/BCJ20200502. Biochem J. 2021. PMID: 33861845 Free PMC article. Review. - Quantitative measurement of phosphoproteome response to osmotic stress in arabidopsis based on Library-Assisted eXtracted Ion Chromatogram (LAXIC).
Xue L, Wang P, Wang L, Renzi E, Radivojac P, Tang H, Arnold R, Zhu JK, Tao WA. Xue L, et al. Mol Cell Proteomics. 2013 Aug;12(8):2354-69. doi: 10.1074/mcp.O113.027284. Epub 2013 May 8. Mol Cell Proteomics. 2013. PMID: 23660473 Free PMC article. - Technical advances in proteomics: new developments in data-independent acquisition.
Hu A, Noble WS, Wolf-Yadlin A. Hu A, et al. F1000Res. 2016 Mar 31;5:F1000 Faculty Rev-419. doi: 10.12688/f1000research.7042.1. eCollection 2016. F1000Res. 2016. PMID: 27092249 Free PMC article. Review. - A self-validating quantitative mass spectrometry method for assessing the accuracy of high-content phosphoproteomic experiments.
Casado P, Cutillas PR. Casado P, et al. Mol Cell Proteomics. 2011 Jan;10(1):M110.003079. doi: 10.1074/mcp.M110.003079. Epub 2010 Oct 24. Mol Cell Proteomics. 2011. PMID: 20972267 Free PMC article.
References
- Hunter T. Cell. 2000;100:113–127. - PubMed
- Pawson T, Scott JD. Trends Biochem Sci. 2005;30:286–290. - PubMed
- Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S. Science. 2002;298:1912–1934. - PubMed
- Songyang Z, Blechner S, Hoagland N, Hoekstra MF, Piwnica-Worms H, Cantley LC. Curr Biol. 1994;4:973–982. - PubMed
- Hutti JE, Jarrell ET, Chang JD, Abbott DW, Storz P, Toker A, Cantley LC, Turk BE. Nat Methods. 2004;1:27–29. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases