The specialized cytosolic J-protein, Jjj1, functions in 60S ribosomal subunit biogenesis - PubMed (original) (raw)
The specialized cytosolic J-protein, Jjj1, functions in 60S ribosomal subunit biogenesis
Alison E Meyer et al. Proc Natl Acad Sci U S A. 2007.
Abstract
J-proteins and Hsp70 chaperones function together in diverse cellular processes. We identified a cytosolic J-protein, Jjj1, of Saccharomyces cerevisiae that is associated with 60S ribosomal particles. Unlike Zuo1, a 60S subunit-associated J-protein that is a component of the chaperone machinery that binds nascent polypeptide chains upon their exit from the ribosome, Jjj1 plays a role in ribosome biogenesis. Cells lacking Jjj1 have phenotypes very similar to those lacking Rei1, a ribosome biogenesis factor associated with pre-60S ribosomal particles in the cytosol. Jjj1 stimulated the ATPase activity of the general cytosolic Hsp70 Ssa, but not Ssb, Zuo1's ribosome-associated Hsp70 partner. Overexpression of Jjj1, which is normally approximately 40-fold less abundant than Zuo1, can partially rescue the phenotypes of cells lacking Zuo1 as well as cells lacking Ssb. Together, these results are consistent with the idea that Jjj1 normally functions with Ssa in a late, cytosolic step of the biogenesis of 60S ribosomal subunits. In addition, because of its ability to bind 60S subunits, we hypothesize that Jjj1, when overexpressed, is able to partially substitute for the Zuo1:Ssb chaperone machinery by recruiting Ssa to the ribosome, facilitating its interaction with nascent polypeptide chains.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
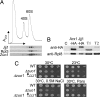
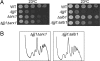
Overexpression of Jjj1, which comigrates with 60S subunits, partially rescues Δ_zuo1_. (A) Lysate from a WT strain was prepared in magnesium-free buffer and separated on a 15–30% sucrose gradient. The OD254 was monitored (Upper). Fractions were collected and analyzed for the presence of Jjj1, Zuo1, and Rpl3 by immunoblotting (Lower). (B) Extracts from cells containing plasmid borne Jjj1-HA or, as a control, genomically integrated Arx1-HA, were incubated with anti-HA antibody and protein A beads. Precipitated proteins were eluted from protein A beads in sample buffer and separated by SDS/PAGE. Total protein extract from Arx-HA cells (T1) and Jjj1-HA cells (T2) as well as immunoprecipitation from an untagged strain (C) were included as controls. Immunoblotting was performed by using antibodies specific for the HA or Rpl8 epitopes. (C) Tenfold serial dilutions of cells were spotted on minimal media plates with indicated additions and incubated at indicated temperatures; pRS415GPD JJJ1 (↑JJJ1); paromomycin (Paro).
Fig. 2.
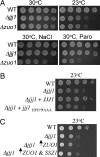
Jjj1 is a J-protein with a function distinct from Zuo1. Approximately equal numbers of cells of the indicated genotypes were diluted 10-fold, spotted on plates, and incubated under conditions indicated on rich media (A), minimal media; pRS316 JJJ1(+JJJ1) or pRS316 jjj1_HPD→AAA (+jjj1_HPD→AAA) (B), and minimal media; pRS416TEF ZUO1 (↑_ZUO1); pRS415GPD SSZ1 (↑_SSZ1) (C).
Fig. 3.
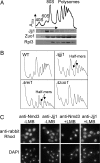
Cells lacking Jjj1, a strictly cytosolic protein, accumulate half-mer ribosomes at 23°C. (A) Ten OD254 units of lysate from WT cells grown at 30°C were centrifuged through a 5–50% sucrose gradient and the OD254 monitored (Upper). Fractions collected were analyzed by immunoblotting for the presence of Jjj1, Zuo1, and Rpl3 (Lower). (B) Ten OD254 units of lysate from the indicated strains grown at 23°C were centrifuged through 7–47% sucrose gradients, and the OD254 was monitored. (C) Log-phase cells of the LMB-sensitive strain AJY1539 were incubated in LMB (0.1 μg/ml) for 30 min and then fixed with formaldehyde and subjected to indirect immunofluorescence microscopy by using antibodies specific for Jjj1 or Nmd3. Localization was then monitored by using anti-rabbit rhodamine-conjugated antibodies. DAPI was used to visualize nuclei.
Fig. 4.
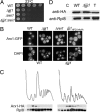
Arx1 fails to recycle in Δ_jjj1_ cells. (A) Approximately equal numbers of cells of the indicated genotypes were diluted 10-fold, spotted onto rich media, and incubated at 23°C. (B) Localization of Arx1-GFP was monitored by fluorescence microscopy in the indicated strains. All cells were grown at 25°C in rich media, except Δ_jjj1_ carrying pRS316 jjj1_HPD→AAA (jjj1HPD→AAA), which was cultured in minimal media lacking uracil. (C) Lysates from 25°C-grown cells containing chromosomally integrated ARX1-HA were centrifuged through 7–47% sucrose gradients and the OD254 monitored (Upper). Fractions were analyzed for the presence of Arx1-HA and Rpl8 by immunoblot analysis (Lower). (D) Extracts from WT or Δ_jjj1 cells containing genomically integrated Arx1-HA were incubated with anti-HA antibody and protein A beads. Precipitated proteins were eluted from protein A beads in sample buffer and separated by SDS/PAGE. Total protein extract (T) as well as immunoprecipitation from an untagged strain (C) were included as controls. Immunoblotting was performed by using antibodies specific for the HA or Rpl8.
Fig. 5.
Deletion of ARX1 or ALB1 suppresses defects of Δ_jjj1_ cells. (A) Approximately equal numbers of cells of the indicated genotypes were diluted 10-fold and spotted on rich media. Plates were incubated at 23°C. (B) Ten OD254 units of lysate from the indicated strains grown at 23°C were centrifuged through 5–50% sucrose gradients and the OD254 was monitored.
Fig. 6.
Jjj1 stimulates the ATPase activity of Ssa1. (A) Ten OD254 units of lysate from the Δ_ssb1_ Δ ssb2 (Δ_ssb1/2_) grown at 23°C were centrifuged through a 5–50% sucrose gradient, and the OD254 was monitored. (B) Approximately equal numbers of cells of the indicated genotypes were diluted 10-fold and spotted on minimal media. Plates were incubated at 23°C. (C) Hsp70–ATP-[32P] complexes were incubated in the presence of various concentrations of Jjj1, Jjj1HPD→AAA, Ydj1, or Zuo1–Ssz1 heterodimer. The rates of hydrolysis of ATP were determined, and fold stimulation over the basal rate was plotted.
Similar articles
- The Hsp40 chaperone Jjj1 is required for the nucleo-cytoplasmic recycling of preribosomal factors in Saccharomyces cerevisiae.
Demoinet E, Jacquier A, Lutfalla G, Fromont-Racine M. Demoinet E, et al. RNA. 2007 Sep;13(9):1570-81. doi: 10.1261/rna.585007. Epub 2007 Jul 24. RNA. 2007. PMID: 17652132 Free PMC article. - Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - The novel ATP-binding cassette protein ARB1 is a shuttling factor that stimulates 40S and 60S ribosome biogenesis.
Dong J, Lai R, Jennings JL, Link AJ, Hinnebusch AG. Dong J, et al. Mol Cell Biol. 2005 Nov;25(22):9859-73. doi: 10.1128/MCB.25.22.9859-9873.2005. Mol Cell Biol. 2005. PMID: 16260602 Free PMC article. - Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.
Ryan R, Hill S. Ryan R, et al. Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article. - Honey as a topical treatment for wounds.
Jull AB, Cullum N, Dumville JC, Westby MJ, Deshpande S, Walker N. Jull AB, et al. Cochrane Database Syst Rev. 2015 Mar 6;2015(3):CD005083. doi: 10.1002/14651858.CD005083.pub4. Cochrane Database Syst Rev. 2015. PMID: 25742878 Free PMC article. Review.
Cited by
- Cryo-EM structures of Arx1 and maturation factors Rei1 and Jjj1 bound to the 60S ribosomal subunit.
Greber BJ, Boehringer D, Montellese C, Ban N. Greber BJ, et al. Nat Struct Mol Biol. 2012 Dec;19(12):1228-33. doi: 10.1038/nsmb.2425. Epub 2012 Nov 11. Nat Struct Mol Biol. 2012. PMID: 23142985 - A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes.
Koplin A, Preissler S, Ilina Y, Koch M, Scior A, Erhardt M, Deuerling E. Koplin A, et al. J Cell Biol. 2010 Apr 5;189(1):57-68. doi: 10.1083/jcb.200910074. J Cell Biol. 2010. PMID: 20368618 Free PMC article. - Role and dynamics of the ribosomal protein P0 and its related trans-acting factor Mrt4 during ribosome assembly in Saccharomyces cerevisiae.
Rodríguez-Mateos M, García-Gómez JJ, Francisco-Velilla R, Remacha M, de la Cruz J, Ballesta JP. Rodríguez-Mateos M, et al. Nucleic Acids Res. 2009 Dec;37(22):7519-32. doi: 10.1093/nar/gkp806. Nucleic Acids Res. 2009. PMID: 19789271 Free PMC article. - Mechanistic insight into eukaryotic 60S ribosomal subunit biogenesis by cryo-electron microscopy.
Greber BJ. Greber BJ. RNA. 2016 Nov;22(11):1643-1662. doi: 10.1261/rna.057927.116. RNA. 2016. PMID: 27875256 Free PMC article. Review. - Synthetic lethal interactions in yeast reveal functional roles of J protein co-chaperones.
Gillies AT, Taylor R, Gestwicki JE. Gillies AT, et al. Mol Biosyst. 2012 Nov;8(11):2901-8. doi: 10.1039/c2mb25248a. Mol Biosyst. 2012. PMID: 22851130 Free PMC article.
References
- Craig EA, Huang P, Aron R, Andrew A. Rev Physiol Biochem Pharmacol. 2006;156:1–21. - PubMed
- Bukau B, Weissman J, Horwich A. Cell. 2006;125:443–451. - PubMed
- Craig EA, Huang P. In: Protein Folding Handbook. Buchner J, Kiefhaber T, editors. Vol 4. Weinheim, Germany: Wiley; 2005. pp. 490–515.
Publication types
MeSH terms
Substances
Grants and funding
- GM31107/GM/NIGMS NIH HHS/United States
- GM53655/GM/NIGMS NIH HHS/United States
- R01 GM031107/GM/NIGMS NIH HHS/United States
- R01 GM053655/GM/NIGMS NIH HHS/United States
- R37 GM031107/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials