High-density yeast-tiling array reveals previously undiscovered introns and extensive regulation of meiotic splicing - PubMed (original) (raw)
High-density yeast-tiling array reveals previously undiscovered introns and extensive regulation of meiotic splicing
Kara Juneau et al. Proc Natl Acad Sci U S A. 2007.
Abstract
Knowing gene structure is vital to understanding gene function, and accurate genome annotation is essential for understanding cellular function. To this end, we have developed a genome-wide assay for mapping introns in Saccharomyces cerevisiae. Using high-density tiling arrays, we compared wild-type yeast to a mutant deficient for intron degradation. Our method identified 76% of the known introns, confirmed 18 previously predicted introns, and revealed 9 formerly undiscovered introns. Furthermore, we discovered that all 13 meiosis-specific intronic yeast genes undergo regulated splicing, which provides posttranscriptional regulation of the genes involved in yeast cell differentiation. Moreover, we found that approximately 16% of intronic genes in yeast are incompletely spliced during exponential growth in rich medium, which suggests that meiosis is not the only biological process regulated by splicing. Our tiling-array assay provides a snapshot of the spliced transcriptome in yeast. This robust methodology can be used to explore environmentally distinct splicing responses and should be readily adaptable to the study of other organisms, including humans.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
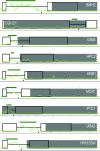
Fig. 1.
Nine introns were discovered by using a high-density tiling array. Intronless gene annotations from the SGD are shown as solid gray boxes (8, 9). For large genes, the uninformative 3′ ends are not shown; truncations are indicated by a jagged edge on the gray boxes. Newly annotated introns are depicted as thin black lines flanked by open boxes, which designate the position of high-quality exonic sequence. Light-green lines graph the intensity that results when wild-type data are subtracted from _dbr1_Δ/_dbr1_Δ data; peaks are indicative of increasing levels of signal from intron lariats. Dark-green bars show intronic intervals automatically identified by the software. Black arrows designate the position of conserved branchpoint sequences. Genomic distances are specified along the thin black line beneath the gene annotations; large tick marks demarcate 100 bp. Gene names are specified in white lettering.
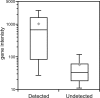
Fig. 2.
Undetected introns are predominantly located in minimally expressed genes. The y axis specifies gene intensity, which is calculated by averaging probe intensities across the exons of each i-gene. ‘Detected’ introns were identified by using our array-based methodology; ‘Undetected’ introns were not identified. On the bar-and-whiskers diagram, the lowest, second-lowest, middle, second-highest, and highest lines represent the 10th percentile, 25th percentile, median, 75th percentile, and 90th percentile, respectively. The gray diamonds designate the mean.
Fig. 3.
Meiosis-specific genes are regulated posttranscriptionally by splicing. PCR products, from primers that surround intronic sequences in 16 meiotic genes (designated above), were separated by using gel electrophoresis on an agarose gel stained with SYBR green. The image color has been inverted for clarity; bands of DNA appear dark on a light background. For each gene, five PCRs were carried out under identical conditions by using five different SK1-derived templates: total RNA (RNA) as a control for genomic contamination, genomic DNA (gDNA), cDNA from cells grown in rich media after 0 hours of sporulation (0h), cDNA from cells harvested after 4 (4h) or 8 (8h) hours of sporulation. The larger PCR products result from genomic DNA in the gDNA lanes or unspliced pre-mRNA in the 0h, 4h, and 8h lanes. The smaller products result from spliced mRNA. The marker (m) is a 50-bp marker. All 13 meiosis-specific genes perform regulated splicing. Only GLC7, TUB1, and TUB3, which have important cellular functions outside of meiosis, are completely spliced in rich media and do not display regulated splicing activity.
Similar articles
- Genome-wide analysis of pre-mRNA splicing: intron features govern the requirement for the second-step factor, Prp17 in Saccharomyces cerevisiae and Schizosaccharomyces pombe.
Sapra AK, Arava Y, Khandelia P, Vijayraghavan U. Sapra AK, et al. J Biol Chem. 2004 Dec 10;279(50):52437-46. doi: 10.1074/jbc.M408815200. Epub 2004 Sep 27. J Biol Chem. 2004. PMID: 15452114 - Genome-wide identification of spliced introns using a tiling microarray.
Zhang Z, Hesselberth JR, Fields S. Zhang Z, et al. Genome Res. 2007 Apr;17(4):503-9. doi: 10.1101/gr.6049107. Epub 2007 Mar 9. Genome Res. 2007. PMID: 17351133 Free PMC article. - The quest for a message: budding yeast, a model organism to study the control of pre-mRNA splicing.
Meyer M, Vilardell J. Meyer M, et al. Brief Funct Genomic Proteomic. 2009 Jan;8(1):60-7. doi: 10.1093/bfgp/elp002. Epub 2009 Mar 11. Brief Funct Genomic Proteomic. 2009. PMID: 19279072 Review. - Meiosis-specific RNA splicing in yeast.
Engebrecht JA, Voelkel-Meiman K, Roeder GS. Engebrecht JA, et al. Cell. 1991 Sep 20;66(6):1257-68. doi: 10.1016/0092-8674(91)90047-3. Cell. 1991. PMID: 1840507 - Functional annotations for the Saccharomyces cerevisiae genome: the knowns and the known unknowns.
Christie KR, Hong EL, Cherry JM. Christie KR, et al. Trends Microbiol. 2009 Jul;17(7):286-94. doi: 10.1016/j.tim.2009.04.005. Epub 2009 Jul 2. Trends Microbiol. 2009. PMID: 19577472 Free PMC article. Review.
Cited by
- Effects of the Paf1 complex and histone modifications on snoRNA 3'-end formation reveal broad and locus-specific regulation.
Tomson BN, Crisucci EM, Heisler LE, Gebbia M, Nislow C, Arndt KM. Tomson BN, et al. Mol Cell Biol. 2013 Jan;33(1):170-82. doi: 10.1128/MCB.01233-12. Epub 2012 Oct 29. Mol Cell Biol. 2013. PMID: 23109428 Free PMC article. - Alternative splicing acting as a bridge in evolution.
Zhou K, Salamov A, Kuo A, Aerts AL, Kong X, Grigoriev IV. Zhou K, et al. Stem Cell Investig. 2015 Oct 30;2:19. doi: 10.3978/j.issn.2306-9759.2015.10.01. eCollection 2015. Stem Cell Investig. 2015. PMID: 27358887 Free PMC article. - RNA secondary structure mediates alternative 3'ss selection in Saccharomyces cerevisiae.
Plass M, Codony-Servat C, Ferreira PG, Vilardell J, Eyras E. Plass M, et al. RNA. 2012 Jun;18(6):1103-15. doi: 10.1261/rna.030767.111. Epub 2012 Apr 26. RNA. 2012. PMID: 22539526 Free PMC article. - Population genomics of intron splicing in 38 Saccharomyces cerevisiae genome sequences.
Skelly DA, Ronald J, Connelly CF, Akey JM. Skelly DA, et al. Genome Biol Evol. 2009 Nov 17;1:466-78. doi: 10.1093/gbe/evp046. Genome Biol Evol. 2009. PMID: 20333215 Free PMC article. - Introns regulate gene expression in Cryptococcus neoformans in a Pab2p dependent pathway.
Goebels C, Thonn A, Gonzalez-Hilarion S, Rolland O, Moyrand F, Beilharz TH, Janbon G. Goebels C, et al. PLoS Genet. 2013;9(8):e1003686. doi: 10.1371/journal.pgen.1003686. Epub 2013 Aug 15. PLoS Genet. 2013. PMID: 23966870 Free PMC article.
References
- Burge CB, Tuschl T, Sharp PA. In: Cold Spring Harbor Monograph Series. Gesteland RF, Cech T, Atkins JF, editors. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1999. pp. 525–560.
- Engebrecht JA, Voelkel-Meiman K, Roeder GS. Cell. 1991;66:1257–1268. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases