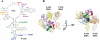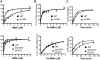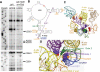Optimization of ribosome structure and function by rRNA base modification - PubMed (original) (raw)
Optimization of ribosome structure and function by rRNA base modification
Jennifer L Baxter-Roshek et al. PLoS One. 2007.
Abstract
Background: Translating mRNA sequences into functional proteins is a fundamental process necessary for the viability of organisms throughout all kingdoms of life. The ribosome carries out this process with a delicate balance between speed and accuracy. This work investigates how ribosome structure and function are affected by rRNA base modification. The prevailing view is that rRNA base modifications serve to fine tune ribosome structure and function.
Methodology/principal findings: To test this hypothesis, yeast strains deficient in rRNA modifications in the ribosomal peptidyltransferase center were monitored for changes in and translational fidelity. These studies revealed allele-specific sensitivity to translational inhibitors, changes in reading frame maintenance, nonsense suppression and aa-tRNA selection. Ribosomes isolated from two mutants with the most pronounced phenotypic changes had increased affinities for aa-tRNA, and surprisingly, increased rates of peptidyltransfer as monitored by the puromycin assay. rRNA chemical analyses of one of these mutants identified structural changes in five specific bases associated with the ribosomal A-site.
Conclusions/significance: Together, the data suggest that modification of these bases fine tune the structure of the A-site region of the large subunit so as to assure correct positioning of critical rRNA bases involved in aa-tRNA accommodation into the PTC, of the eEF-1A.aa-tRNA.GTP ternary complex with the GTPase associated center, and of the aa-tRNA in the A-site. These findings represent a direct demonstration in support of the prevailing hypothesis that rRNA modifications serve to optimize rRNA structure for production of accurate and efficient ribosomes.
Conflict of interest statement
Competing Interests: The authors have declared that no competing interests exist.
Figures
Figure 1. 25S rRNA in the peptiptidyl transferase center of yeast.
(A) Secondary structure of yeast 25S rRNA in the PTC. snoRNAs targeted for this study are indicated along with the residues they modify. Ψ – pseudouridylated residue; Nm – 2′-_O_-ribose methylated residue. Helices are numbered in black. (B) Three dimensional representation of the E. coli PTC . Modified residues are labeled by the colors indicated in panel A. Left: view into the PTC from the top of the LSU, _righ_t: 90° rotation of Left. Helices and tRNAs are labeled.
Figure 2. Sensitivity of rRNA base modification mutants to Translational Inhibitors.
Mutant and isogenic wild-type yeast strains were spotted as ten fold dilutions from 105 to 101 CFU onto YPAD media containing 20 µg/ml anisomycin or sparsomycin. Cells were incubated for 3 days at 30°C, and growth was monitored as compared to growth on plates in the absence of drug. Each strain and drug was assayed at least twice.
Figure 3. Many of the rRNA base modification mutants have M1 virus propagation defects.
Yeast rRNA modification mutants were tested for their ability to maintain the L-A and M1 viruses. (A) Mutant and isogenic wild-type yeast strains were spotted onto YPAD plates, and allowed to grow at 30°C, and then replica plated to a seeded lawn of 5X47 indicator cells. Plates were incubated at room temperature for 3–5 days until a zone of inhibition was clearly visible for wild-type cells. (B) Total RNAs were extracted from mutant and isogenic wild-type yeast strains and digested with RNase A under high salt conditions. The resulting double-stranded RNA was separated on a 1% agarose gel and visualized with ethidium bromide. L-A and M1 dsRNAs are indicated. The image was inverted for clarity.
Figure 4. Ribosome biochemistry.
Mutant strain snr46 Δ and the isogenic wild-type are shown in the top row, and mutant strain spb1DA/snr52 Δ and the isogenic wild-type are show in the bottom row. Error bars represent standard error for all graphs. A. [14C]Phe-tRNA binding to the A-site of the ribosome. One site binding curves of bound tRNA as analyzed using GraphPad Prism software. Data are reported as a percentage of the total tRNA bound. B. Ac-[14C]Phe-tRNA binding to the P-site of the ribosome. One site binding curves of bound tRNA as analyzed using GraphPad Prism software. Data are reported as a percentage of the total tRNA bound. C. Peptidyltransfer. Timecourse assays of peptidyltransferase activities as measured by the puromycin reaction.
Figure 5. 25S rRNA structure probing analysis in spb1DA/snr52 Δ mutant ribosomes.
(A) Puromycin treated ribosomes isolated from isogenic wild-type and spb1DA/snr52 Δ mutant strains were used for in vitro chemical probing of the structure around the peptidyltransferase center of the ribosome, specifically helices 89-93. Reactions were performed in triplicate, representative autoradiographs are shown. U - untreated; C - CMCT; D – DMS; K – Kethoxal. Residues with changes in banding pattern labeled. Strongly modified bases at positions U2923, C2843, and C2851 are indicated, as are the more weakly deprotected A2932 and A2933. The increased intensity corresponding to U2845 (marked by *) is not DMS-specific. (B–D). Locations of residues demonstrating strong changes in protection patterns mapped to LSU rRNA structures. Um2920 and Gm2921 are indicated and color coded. Bases with altered protection patterns are circled in orange. aa-tRNA accommodation ‘gate’ bases are indicated with purple (gate 1) and green (gate 2). The “catalytic” A base (equivalent to E. coli 23S rRNA A2451) is incicated with red. Individual LSU helices are numbered and color coded. (B) Secondary structure of yeast 25S rRNA around the PTC. (C) Three dimensional representation of rRNA bases of interest mapped onto the E. coli PTC . Arrow represents the path the 3′ end of the aa-tRNA travels when entering the A-site. (D) Rotation and zoom in of Panel C. Shows the path into the A-site from the aa-tRNA perspective. Residues demonstrating changes in protection patterns are labeled in orange. Locations of modified bases and A-site ‘gate’ residues are labeled.
Similar articles
- Mutations of highly conserved bases in the peptidyltransferase center induce compensatory rearrangements in yeast ribosomes.
Rakauskaite R, Dinman JD. Rakauskaite R, et al. RNA. 2011 May;17(5):855-64. doi: 10.1261/rna.2593211. Epub 2011 Mar 25. RNA. 2011. PMID: 21441349 Free PMC article. - An arc of unpaired "hinge bases" facilitates information exchange among functional centers of the ribosome.
Rakauskaite R, Dinman JD. Rakauskaite R, et al. Mol Cell Biol. 2006 Dec;26(23):8992-9002. doi: 10.1128/MCB.01311-06. Epub 2006 Sep 25. Mol Cell Biol. 2006. PMID: 17000775 Free PMC article. - rRNA mutants in the yeast peptidyltransferase center reveal allosteric information networks and mechanisms of drug resistance.
Rakauskaite R, Dinman JD. Rakauskaite R, et al. Nucleic Acids Res. 2008 Mar;36(5):1497-507. doi: 10.1093/nar/gkm1179. Epub 2008 Jan 18. Nucleic Acids Res. 2008. PMID: 18203742 Free PMC article. - Structural dynamics of ribosomal RNA during decoding on the ribosome.
Rodnina MV, Daviter T, Gromadski K, Wintermeyer W. Rodnina MV, et al. Biochimie. 2002 Aug;84(8):745-54. doi: 10.1016/s0300-9084(02)01409-8. Biochimie. 2002. PMID: 12457562 Review. - [Mechanism of peptide bond formation on the ribosome--controversions].
Bakowska-Zywicka K, Tyczewska A, Twardowski T. Bakowska-Zywicka K, et al. Postepy Biochem. 2006;52(2):166-72. Postepy Biochem. 2006. PMID: 17078506 Review. Polish.
Cited by
- Evaluating the reproducibility of quantifying modified nucleosides from ribonucleic acids by LC-UV-MS.
Russell SP, Limbach PA. Russell SP, et al. J Chromatogr B Analyt Technol Biomed Life Sci. 2013 Apr 1;923-924:74-82. doi: 10.1016/j.jchromb.2013.02.010. J Chromatogr B Analyt Technol Biomed Life Sci. 2013. PMID: 23500350 Free PMC article. - Yeast Rrp8p, a novel methyltransferase responsible for m1A 645 base modification of 25S rRNA.
Peifer C, Sharma S, Watzinger P, Lamberth S, Kötter P, Entian KD. Peifer C, et al. Nucleic Acids Res. 2013 Jan;41(2):1151-63. doi: 10.1093/nar/gks1102. Epub 2012 Nov 23. Nucleic Acids Res. 2013. PMID: 23180764 Free PMC article. - Deciphering the epitranscriptome: A green perspective.
Burgess A, David R, Searle IR. Burgess A, et al. J Integr Plant Biol. 2016 Oct;58(10):822-835. doi: 10.1111/jipb.12483. Epub 2016 Jun 20. J Integr Plant Biol. 2016. PMID: 27172004 Free PMC article. Review. - Ribosome biogenesis factor Tsr3 is the aminocarboxypropyl transferase responsible for 18S rRNA hypermodification in yeast and humans.
Meyer B, Wurm JP, Sharma S, Immer C, Pogoryelov D, Kötter P, Lafontaine DL, Wöhnert J, Entian KD. Meyer B, et al. Nucleic Acids Res. 2016 May 19;44(9):4304-16. doi: 10.1093/nar/gkw244. Epub 2016 Apr 15. Nucleic Acids Res. 2016. PMID: 27084949 Free PMC article. - Deciphering the Role of the Non-Coding Genome in Regulating Gene-Diet Interactions.
Law PP, Holland ML. Law PP, et al. Nutrients. 2018 Nov 27;10(12):1831. doi: 10.3390/nu10121831. Nutrients. 2018. PMID: 30486341 Free PMC article. Review.
References
- Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. - PubMed
- Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, et al. Crystal Structure of the Ribosome at 5.5 A Resolution. Science. 2001;292:883–896. - PubMed
- Decatur WA, Fournier MJ. rRNA modifications and ribosome function. Trends Biochem Sci. 2002;27:344–351. - PubMed
- Ofengand J, Bakin A, Wrzesinski J, Nurse K, Lane BG. The pseudouridine residues of ribosomal RNA. Biochem Cell Biol. 1995;73:915–924. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous