Diagnosis of the small round blue cell tumors using multiplex polymerase chain reaction - PubMed (original) (raw)
Comparative Study
Diagnosis of the small round blue cell tumors using multiplex polymerase chain reaction
Qing-Rong Chen et al. J Mol Diagn. 2007 Feb.
Abstract
The small round blue cell tumors of childhood, which include neuroblastoma, rhabdomyosarcoma, non-Hodgkin's lymphoma, and the Ewing's family of tumors, are so called because of their similar appearance on routine histology. Using cDNA microarray gene expression profiles and artificial neural networks (ANNs), we previously identified 93 genes capable of diagnosing these cancers. Using a subset of these, together with some additional genes (total 39), we developed a multiplex polymerase chain reaction (PCR) assay to diagnose these cancer types. Blinded testing of 96 new samples (26 Ewing's family of tumors, 29 rhabdomyosarcomas, 24 neuroblastomas, and 17 lymphomas) using ANNs in a complete leave-one-out analysis demonstrated that all except one sample were accurately diagnosed as their respective category. Moreover, using an ANN-based gene minimization strategy in a separate analysis, we found that the top 31 genes could correctly diagnose all 96 tumors. Our results suggest that this molecular test based on a multiplex PCR reaction may assist the physician in the rapid confirmation of the diagnosis of these cancers.
Figures
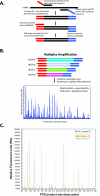
Figure 1
The schematic illustration of multiplex RT-PCR assay. A: The multiplex RT-PCR involves two stages: the first stage includes reverse transcription and amplification using chimeric primers, and the second stage converts to the use of a single pair of universal primers during amplification (see Materials and Methods for the primer design). B: The amplicons obtained from multiplex amplification were then analyzed using fluorescence capillary electrophoresis. The peak location represents the gene identity, and the peak area represents gene expression level. C: The comparative chromatograms of four different categories of tumor samples from one multiplex assay. Blue, Lymph-13; yellow, EWS-T-4; red, RMS-A-18; and green, NB-20.
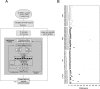
Figure 2
The artificial neural network. A: Workflow for a complete leave-one-out ANN analysis. Multiplex RT-PCR analysis using 40 genes was performed on tumors from 96 pediatric cancer patients (26 EWS, 29 RMSs, 17 lymphomas, and 24 NBs). One sample was left out as an independent test sample, and the ANNs were trained using the remaining 95 samples. ANN training scheme (gray box). 1, All samples were randomly partitioned into three groups. 2, One of the three groups (containing 32 samples) was selected as a validation set, whereas the remaining two groups (63 samples) were used to train the network. 3 and 4, The training weights were iteratively adjusted for 100 cycles (epochs). 5, The ANN output (0 to 1) for each of four classes (EWS, RMS, NB, and lymphoma) was calculated for each sample in the validation set. 6, A different validation set was selected from the same partitioning in 1, and the remaining two groups were used for training. Steps 2 through 6 were repeated until each of the three groups from 1 had been used as a validation set exactly one time. 7, The samples were randomly repartitioned into three new groups, and steps 2 through 6 were repeated. Sample partitioning was performed 100 times in total. Thus, steps 1 through 6 were repeated 100 times. Three hundred ANN models were thus trained and were used to predict the left-out test sample. This scheme was repeated for each left-out test sample. B: Classification of the samples from a leave-one-out ANN analysis. A sample is classified to a cancer category according to its highest committee vote (average of all ANN outputs; Table 1). Plotted is the distance for each sample from its committee vote to the ideal vote for that category (for example, for EWS, it is EWS = 1, RMS = NB = Lymph = 0). The perfectly classified sample would be plotted with a distance of 0. The histological diagnosis of four different cancer categories was displayed in shape as diamond for EWS, square for RMS, triangle for NB, and circle for lymphoma. All samples were correctly classified except one RMS sample, which was misclassified as EWS.
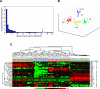
Figure 3
Hierarchical clustering and multidimensional scaling analysis. A: Gene minimization plot for ANN prediction. All of 39 genes were used for the analysis of 96 samples. ANNs were first trained using 96 samples, and 39 genes were ranked according to their importance to the ANN prediction. Red arrow marked the position of 31 genes. B: Multidimensional scaling analysis using 31 top-ranked genes. Three dimensions of the multidimensional scaling plot are shown. EWSs are depicted as yellow circles, RMS as red, NB as green, and lymphoma as blue. The samples clustered closely according to the four different cancer categories. C: Hierarchical clustering of all 96 samples and 31 top-ranked genes. Each row represents a gene, and each column, a separate sample. A pseudocolored representation of the ratio (log2-transformed and _z_-scored across the samples) is shown. On the right are the gene symbols of 31 genes as well as the ANN gene rank.
Similar articles
- Role of immunocytochemistry and DNA flow cytometry in the fine-needle aspiration diagnosis of malignant small round-cell tumors.
Brahmi U, Rajwanshi A, Joshi K, Ganguly NK, Vohra H, Gupta SK, Dey P. Brahmi U, et al. Diagn Cytopathol. 2001 Apr;24(4):233-9. doi: 10.1002/dc.1050. Diagn Cytopathol. 2001. PMID: 11285617 - Reverse transcriptase-polymerase chain reaction as an ancillary molecular technique in the diagnosis of small blue round cell tumors by fine-needle aspiration cytology.
Gautam U, Srinivasan R, Rajwanshi A, Bansal D, Marwaha RK, Vasishtha RK. Gautam U, et al. Am J Clin Pathol. 2010 Apr;133(4):633-45. doi: 10.1309/AJCPPJJ0PY4XZOEC. Am J Clin Pathol. 2010. PMID: 20231617 - Multiplex RT-PCR assay for the differential diagnosis of alveolar rhabdomyosarcoma and Ewing's sarcoma.
Downing JR, Khandekar A, Shurtleff SA, Head DR, Parham DM, Webber BL, Pappo AS, Hulshof MG, Conn WP, Shapiro DN. Downing JR, et al. Am J Pathol. 1995 Mar;146(3):626-34. Am J Pathol. 1995. PMID: 7887445 Free PMC article. - Cytogenetic and pathologic aspects of Ewing's sarcoma and neuroectodermal tumors.
Stephenson CF, Bridge JA, Sandberg AA. Stephenson CF, et al. Hum Pathol. 1992 Nov;23(11):1270-7. doi: 10.1016/0046-8177(92)90295-e. Hum Pathol. 1992. PMID: 1330877 Review. - Recent advances in the molecular biology, diagnosis and novel therapies for various small blue cell tumors.
Pisick E, Skarin AT, Salgia R. Pisick E, et al. Anticancer Res. 2003 Jul-Aug;23(4):3379-96. Anticancer Res. 2003. PMID: 12926079 Review.
Cited by
- A Nonpediatric Extraosseous Ewing Sarcoma of the Pancreas: Differential Diagnosis and Therapeutic Strategies.
Yohannan B, Feldman M. Yohannan B, et al. Case Rep Oncol Med. 2020 Jan 30;2020:2792750. doi: 10.1155/2020/2792750. eCollection 2020. Case Rep Oncol Med. 2020. PMID: 32082662 Free PMC article. - A novel next generation sequencing approach to improve sarcoma diagnosis.
McConnell L, Houghton O, Stewart P, Gazdova J, Srivastava S, Kim C, Catherwood M, Strobl A, Flanagan AM, Oniscu A, Kroeze LI, Groenen P, Taniere P, Salto-Tellez M, Gonzalez D. McConnell L, et al. Mod Pathol. 2020 Jul;33(7):1350-1359. doi: 10.1038/s41379-020-0488-1. Epub 2020 Feb 11. Mod Pathol. 2020. PMID: 32047232 - Development and validation of a robust multiplex serological assay to quantify antibodies specific to pertussis antigens.
Rajam G, Carlone G, Kim E, Choi J, Paulos S, Park S, Jeyachandran A, Gorantla Y, Wong E, Sabnis A, Browning P, Desai R, Quinn CP, Schiffer J. Rajam G, et al. Biologicals. 2019 Jan;57:9-20. doi: 10.1016/j.biologicals.2018.11.001. Epub 2018 Nov 18. Biologicals. 2019. PMID: 30458978 Free PMC article. - Multi-parameter gene expression profiling of peripheral blood for early detection of hepatocellular carcinoma.
Xie H, Xue YQ, Liu P, Zhang PJ, Tian ST, Yang Z, Guo Z, Wang HM. Xie H, et al. World J Gastroenterol. 2018 Jan 21;24(3):371-378. doi: 10.3748/wjg.v24.i3.371. World J Gastroenterol. 2018. PMID: 29391759 Free PMC article. - Prostatic sarcoma of the Ewing family in a 33-year-old male - A case report and review of the literature.
Esch L, Barski D, Bug R, Otto T. Esch L, et al. Asian J Urol. 2016 Apr;3(2):103-106. doi: 10.1016/j.ajur.2015.11.007. Epub 2015 Dec 19. Asian J Urol. 2016. PMID: 29264173 Free PMC article.
References
- Triche TJ, Askin FB. Neuroblastoma and the differential diagnosis of small-, round-, blue-cell tumors. Hum Pathol. 1983;14:569–595. - PubMed
- McManus AP, Gusterson BA, Pinkerton CR, Shipley JM. The molecular pathology of small round-cell tumours: relevance to diagnosis, prognosis, and classification. J Pathol. 1996;178:116–121. - PubMed
- de Kok JB, Roelofs RW, Giesendorf BA, Pennings JL, Waas ET, Feuth T, Swinkels DW, Span PN. Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab Invest. 2005;85:154–159. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical