Human genetics of infectious diseases: a unified theory - PubMed (original) (raw)
Review
Human genetics of infectious diseases: a unified theory
Jean-Laurent Casanova et al. EMBO J. 2007.
Abstract
Since the early 1950s, the dominant paradigm in the human genetics of infectious diseases postulates that rare monogenic immunodeficiencies confer vulnerability to multiple infectious diseases (one gene, multiple infections), whereas common infections are associated with the polygenic inheritance of multiple susceptibility genes (one infection, multiple genes). Recent studies, since 1996 in particular, have challenged this view. A newly recognised group of primary immunodeficiencies predisposing the individual to a principal or single type of infection is emerging. In parallel, several common infections have been shown to reflect the inheritance of one major susceptibility gene, at least in some populations. This novel causal relationship (one gene, one infection) blurs the distinction between patient-based Mendelian genetics and population-based complex genetics, and provides a unified conceptual frame for exploring the molecular genetic basis of infectious diseases in humans.
Figures
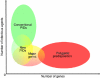
Figure 1
Human genetics of infectious diseases. The spectrum of genetic predisposition to infectious diseases in human patients is represented, according to the number of genes involved (_x_-axis) and the number of infections (_y_-axis). The dominant view in human genetics of infectious diseases postulates that rare, ‘conventional', monogenic primary immunodeficiencies (PIDs, in green) predispose the individual to numerous infections (one gene, multiple infections), whereas common infectious diseases are associated with polygenic inheritance (in red) of numerous susceptibility genes (one infection, multiple genes). Novel monogenic PIDs (in yellow/green) predispose the individual to a principal or single type of infection. Major genes (in yellow/red) exert a nearly Mendelian impact at the population level and largely account for common infectious diseases in some individuals. The recent discovery of such human genes conferring vulnerability or resistance to a specific infection at the individual level (one gene, one infection) bridges the gap between the two classical fields of conventional PIDs and polygenic inheritance, as defined in the 50 s. As an example, genetic predisposition to tuberculosis, which was considered to be purely polygenic, was recently shown to reflect both new PID and major gene effects, at least in some patients (see text for details). Overall, these observations provide experimental support for a continuous spectrum of predisposition and a unified theory of the human genetics of infectious diseases.
Similar articles
- The genomics and genetics of human infectious disease susceptibility.
Hill AV. Hill AV. Annu Rev Genomics Hum Genet. 2001;2:373-400. doi: 10.1146/annurev.genom.2.1.373. Annu Rev Genomics Hum Genet. 2001. PMID: 11701655 Review. - Human genetic susceptibility to intracellular pathogens.
Vannberg FO, Chapman SJ, Hill AV. Vannberg FO, et al. Immunol Rev. 2011 Mar;240(1):105-16. doi: 10.1111/j.1600-065X.2010.00996.x. Immunol Rev. 2011. PMID: 21349089 Review. - Human genetics of infectious diseases: between proof of principle and paradigm.
Alcaïs A, Abel L, Casanova JL. Alcaïs A, et al. J Clin Invest. 2009 Sep;119(9):2506-14. doi: 10.1172/JCI38111. J Clin Invest. 2009. PMID: 19729848 Free PMC article. Review. - Genes that permit or prevent infections.
Thurmon TF. Thurmon TF. J La State Med Soc. 2003 Mar-Apr;155(2):104-7. J La State Med Soc. 2003. PMID: 12778995 Review. - Genetic predisposition to infectious pathogens: a review of less familiar variants.
Somech R, Amariglio N, Spirer Z, Rechavi G. Somech R, et al. Pediatr Infect Dis J. 2003 May;22(5):457-61. doi: 10.1097/01.inf.0000068205.82627.55. Pediatr Infect Dis J. 2003. PMID: 12792391 Review.
Cited by
- Polygenic plague resistance in the great gerbil uncovered by population sequencing.
Nilsson P, Ravinet M, Cui Y, Berg PR, Zhang Y, Guo R, Luo T, Song Y, Trucchi E, Hoff SNK, Lv R, Schmid BV, Easterday WR, Jakobsen KS, Stenseth NC, Yang R, Jentoft S. Nilsson P, et al. PNAS Nexus. 2022 Oct 5;1(5):pgac211. doi: 10.1093/pnasnexus/pgac211. eCollection 2022 Nov. PNAS Nexus. 2022. PMID: 36712379 Free PMC article. - Why children with severe bacterial infection die: a population-based study of determinants and consequences of suboptimal care with a special emphasis on methodological issues.
Launay E, Gras-Le Guen C, Martinot A, Assathiany R, Martin E, Blanchais T, Deneux-Tharaux C, Rozé JC, Chalumeau M. Launay E, et al. PLoS One. 2014 Sep 23;9(9):e107286. doi: 10.1371/journal.pone.0107286. eCollection 2014. PLoS One. 2014. PMID: 25247401 Free PMC article. - Inborn errors of immunity: The missing link in infectious diseases susceptibility.
Pham-Huy A. Pham-Huy A. J Assoc Med Microbiol Infect Dis Can. 2019 Jun 17;4(2):51-54. doi: 10.3138/jammi.2018.11.12. eCollection 2019 Jun. J Assoc Med Microbiol Infect Dis Can. 2019. PMID: 36337741 Free PMC article. No abstract available. - Polymorphism of X-linked CD40 ligand gene associated with pulmonary tuberculosis in the Han Chinese population.
Hu CY, Zhang XA, Meyer CG, Thye T, Liu W, Cao WC. Hu CY, et al. Genes Immun. 2015 Sep;16(6):399-404. doi: 10.1038/gene.2015.20. Epub 2015 Jun 4. Genes Immun. 2015. PMID: 26043172 - How to manage children who have come into contact with patients affected by tuberculosis.
Lancella L, Vecchio AL, Chiappini E, Tadolini M, Cirillo D, Tortoli E, de Martino M, Guarino A, Principi N, Villani A, Esposito S, Galli L; Italian Pediatric TB Study Group. Lancella L, et al. J Clin Tuberc Other Mycobact Dis. 2015 Aug 8;1:1-12. doi: 10.1016/j.jctube.2015.07.002. eCollection 2015 Nov. J Clin Tuberc Other Mycobact Dis. 2015. PMID: 31723675 Free PMC article. Review.
References
- Abel L, Casanova JL (2006) Human genetics of infectious diseases: fundamental insights from clinical studies. Semin Immunol 18: 327–329 - PubMed
- Abel L, Vu DL, Oberti J, Nguyen VT, Van VC, Guilloud-Bataille M, Schurr E, Lagrange PH (1995) Complex segregation analysis of leprosy in southern Vietnam. Genet Epidemiol 12: 63–82 - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical