Differential histone modifications mark mouse imprinting control regions during spermatogenesis - PubMed (original) (raw)
Comparative Study
Differential histone modifications mark mouse imprinting control regions during spermatogenesis
Katia Delaval et al. EMBO J. 2007.
Abstract
Only some imprinting control regions (ICRs) acquire their DNA methylation in the male germ line. These imprints are protected against the global demethylation of the sperm genome following fertilisation, and are maintained throughout development. We find that in somatic cells and tissues, DNA methylation at these ICRs is associated with histone H4-lysine-20 and H3-lysine-9 trimethylation. The unmethylated allele, in contrast, has H3-lysine-4 dimethylation and H3 acetylation. These differential modifications are also detected at maternally methylated ICRs, and could be involved in the somatic maintenance of imprints. To explore whether the post-fertilisation protection of imprints relates to events during spermatogenesis, we assayed chromatin at stages preceding the global histone-to-protamine exchange. At these stages, H3-lysine-4 methylation and H3 acetylation are enriched at maternally methylated ICRs, but are absent at paternally methylated ICRs. H4 acetylation is enriched at all regions analysed. Thus, paternally and maternally methylated ICRs carry different histone modifications during the stages preceding the global histone-to-protamine exchange. These differences could influence the way ICRs are assembled into specific structures in late spermatogenesis, and may thus influence events after fertilisation.
Figures
Figure 1
The ICRs of the Igf2-H19, Dlk1-Gtl2 and Rasgrf1 domains. Genes are represented as grey boxes; arrows indicate their allelic transcription status. ICRs are shown as filled black rectangles; lollipops indicate their paternal CpG methylation. Thin lines indicate regions analysed by PCR amplification and SSCP electrophoresis in precipitated chromatin fractions. The ICR of the Igf2 and H19 genes is located 2–4 kb upstream of H19 (Thorvaldsen et al, 1998), and has DNA methylation (indicated as a black bar) on the paternal chromosome (P). The ICR of the Dlk1-Gtl2 domain is at 10–15 kb upstream of Gtl2 (Lin et al, 2003). Only part of this >1 Mb domain is depicted. Imprinted expression of Rasgrf1 is controlled by an ICR at 30 kb upstream of the gene (Yoon et al, 2002). In the embryo, the paternal methylation covers a region of about 7 kb (Kobayashi et al, 2006).
Figure 2
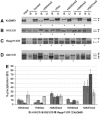
Allelic histone modifications at different ICRs in ES cells. ChIP was performed against H3 lysine-4 dimethylation (H3K4me2), H3 lysine-9+14 acetylation (H3ac), H4 lysine-20 trimethylation (H4K20me3), H3 lysine-9 trimethylation (H3K9me3) or H3 lysine-27 trimethylation (H3K27me3). DNA extracted from antibody-bound (B) and unbound (U) fractions was PCR amplified, followed by electrophoretic detection of SSCPs. In each panel, the left lane shows the input chromatin, followed by a control ChIP (C) without a histone-specific antiserum. For each of the bound fractions, we determined the ratio between the maternal (M) and paternal (P) alleles. Asterisks indicate ChIP samples where, after correction against the allelic ratio in the input chromatin, the allelic ratio was higher than 3 (more than three-fold allelic enrichment). Typically, where only one parental allele is visible in the bound fraction, this corresponds to a >15-fold enrichment. (A) The KvDMR1 regulates imprinted expression along the Kcnq1 domain. The analysed region is at 100 bp from the Kcnq1ot1 transcription start site. (B) The H19 ICR. PCR–SSCP was performed at a region located 2 kb upstream of H19. (C) The ICR of the Rasgrf1 domain. A region at 200 bp from this ICR was analysed by PCR–SSCP. (D) The ICR of the Dlk1-Gtl2 domain.
Figure 3
Allele-specific histone methylation and acetylation in liver. ChIP and allelic discrimination by PCR–SSCP were as for ES cells. Asterisks indicate the lanes where, after correction against the allelic ratio in the input chromatin, the allelic ratios were higher than 3. Typically, where only one parental allele is visible in the bound fraction, this corresponds to a >15-fold enrichment. Shown are ChIP data for KvDMR1 (A), H19 ICR (B), Rasgrf1 ICR (C) and Gtl2 ICR (D). (E) For quantification of histone modifications, ChIP samples were analysed by real-time PCR. Levels of precipitation are shown as the percentage of the input chromatin that was precipitated in the antibody-bound fractions.
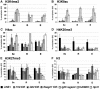
Figure 4
Levels of histone modifications and DNA methylation at different spermatogenic stages. (A) Schematic representation of spermatogenesis and the different spermatogenic stages. The line with arrowhead on the right indicates when DNA methylation is detected at the H19, Gtl2 and Rasgrf1 ICRs and at LINE1 repeats. (B) Total protein fractions corresponding to spermatocytes (Sc), round (samples R1 and R2) and elongating spermatids (E1 and E2) were used for Western blotting. Equal amounts of protein were loaded in the different lanes. Immunostaining was with antisera against different histone modifications, histone H2A and transition protein TP2. Measured band intensities were normalised using H2A, and are presented in the graph to the right. The additional graph display presents data from an independent second experiment. Note that the elongating spermatid fraction 2 corresponds to a later stage than fraction 1, explaining the higher TP2 expression. (C) Analysis of DNA methylation at ICRs in spermatocytes and round and elongating spermatids. Total genomic DNA was digested with the methylation-sensitive enzyme _Cfo_I, followed by real-time PCR amplification across the ICRs of H19, Rasgrf1, Gtl2, KvDMR1 and Igf2r region 2. Also included are the Hprt1 CpG island and LINE1 sequences. Plotted for each locus is the percentage of DNA digested by _Cfo_I. (D) Purification of unfixed chromatin fragments from spermatocytes and round and elongating spermatids. Extracted nuclei were incubated for a short time in a buffer containing MNase. DNA was purified and electrophoresed through a 1% agarose gel, followed by ethidium bromide staining of the gel. In all three preparations, chromatin fragments were 1–7 nucleosomes in length.
Figure 5
H3 modifications distinguish different ICRs in spermatocytes and at postmeiotic stages of spermatogenesis. Levels of H3K4me2 (A), H3K9ac (B), H4ac (C), H4K20me3 (D), H3K27me3 (E) and total H3 (F) were assessed in spermatocytes (Sc), in round spermatids (R), and in elongating spermatids (E), by ChIP followed by quantitative PCR. For each graph, precipitation is shown as the percentage of input chromatin (I) that was brought down by the specific antiserum (B, for bound fraction) at the locus of interest. Values are the average of two to three independent ChIP experiments, and were corrected for background levels of precipitation at the different loci analysed (background precipitation was determined with a non-histone related antiserum and was <3% at all regions analysed).
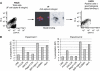
Figure 6
Analysis of H3 K4 methylation in spermatogonia. (A) Purification of type-A spermatogonia from 6 to 7-day-old males. Shown are cells immunostained for integrin alpha-6, purified with magnetic beads. Positively staining cells (bead binding +) are spermatogonia (Shinohara et al, 1999) also visualised by FACS analysis of the sorted fraction. (B) Five hundred thousand spermatogonia were used for ChIP against H3K4me2. Experiments were performed on two independent spermatogonial purifications. Precipitation is shown as the percentage of input chromatin brought down at the locus of interest.
Similar articles
- "I've Spent My Whole Life Striving to Be Normal": Internalized Stigma and Perceived Impact of Diagnosis in Autistic Adults.
Huang Y, Trollor JN, Foley KR, Arnold SRC. Huang Y, et al. Autism Adulthood. 2023 Dec 1;5(4):423-436. doi: 10.1089/aut.2022.0066. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116050 Free PMC article. - Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.
Salisbury L, Baraitser L. Salisbury L, et al. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review. - Using Experience Sampling Methodology to Capture Disclosure Opportunities for Autistic Adults.
Love AMA, Edwards C, Cai RY, Gibbs V. Love AMA, et al. Autism Adulthood. 2023 Dec 1;5(4):389-400. doi: 10.1089/aut.2022.0090. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116059 Free PMC article. - Repetitive Negative Thinking As a Transdiagnostic Prospective Predictor of Depression and Anxiety Symptoms in Neurodiverse First-Semester College Students.
McKenney EE, Brunwasser SM, Richards JK, Day TC, Kofner B, McDonald RG, Williams ZJ, Gillespie-Lynch K, Kang E, Lerner MD, Gotham KO. McKenney EE, et al. Autism Adulthood. 2023 Dec 1;5(4):374-388. doi: 10.1089/aut.2022.0078. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116057 Free PMC article. - Oral dextrose gel to prevent hypoglycaemia in at-risk neonates.
Roberts L, Lin L, Alsweiler J, Edwards T, Liu G, Harding JE. Roberts L, et al. Cochrane Database Syst Rev. 2023 Nov 28;11(11):CD012152. doi: 10.1002/14651858.CD012152.pub4. Cochrane Database Syst Rev. 2023. PMID: 38014716 Free PMC article. Review.
Cited by
- Iterative oxidation by TET1 is required for reprogramming of imprinting control regions and patterning of mouse sperm hypomethylated regions.
Prasasya RD, Caldwell BA, Liu Z, Wu S, Leu NA, Fowler JM, Cincotta SA, Laird DJ, Kohli RM, Bartolomei MS. Prasasya RD, et al. Dev Cell. 2024 Apr 22;59(8):1010-1027.e8. doi: 10.1016/j.devcel.2024.02.012. Epub 2024 Apr 2. Dev Cell. 2024. PMID: 38569549 - Presence of H3K4me3 on Paternally Expressed Genes of the Paternal Genome From Sperm to Implantation.
Ishihara T, Griffith OW, Suzuki S, Renfree MB. Ishihara T, et al. Front Cell Dev Biol. 2022 Mar 10;10:838684. doi: 10.3389/fcell.2022.838684. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35359448 Free PMC article. - Chromatin and Nuclear Dynamics in the Maintenance of Replication Fork Integrity.
Wootton J, Soutoglou E. Wootton J, et al. Front Genet. 2021 Dec 14;12:773426. doi: 10.3389/fgene.2021.773426. eCollection 2021. Front Genet. 2021. PMID: 34970302 Free PMC article. Review. - Deciphering the genetic code of DNA methylation.
Wang M, Ngo V, Wang W. Wang M, et al. Brief Bioinform. 2021 Sep 2;22(5):bbaa424. doi: 10.1093/bib/bbaa424. Brief Bioinform. 2021. PMID: 33432324 Free PMC article. Review. - Linker histone variant H1t is closely associated with repressed repeat-element chromatin domains in pachytene spermatocytes.
Mahadevan IA, Kumar S, Rao MRS. Mahadevan IA, et al. Epigenetics Chromatin. 2020 Mar 4;13(1):9. doi: 10.1186/s13072-020-00335-x. Epigenetics Chromatin. 2020. PMID: 32131873 Free PMC article.
References
- Bellve AR (1993) Purification, culture, and fractionation of spermatogenic cells. Methods Enzymol 225: 84–113 - PubMed
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Jaenisch R, Wagschal A, Feil R, Schreiber SL, Lander ES (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 - PubMed
- Bourc'his D, Xu GL, Lin CS, Bollman B, Bestor TH (2001) Dnmt3L and the establishment of maternal genomic imprints. Science 294: 2536–2539 - PubMed
- Davis TL, Yang GJ, McCarrey JR, Bartolomei MS (2000) The H19 methylation imprint is erased and re-established differentially on the parental alleles during male germ cell development. Hum Mol Genet 9: 2885–2894 - PubMed
- Dean W, Bowden L, Aitchison A, Klose J, Moore T, Meneses JJ, Reik W, Feil R (1998) Altered imprinted gene methylation and expression in completely ES cell-derived mouse fetuses: association with aberrant phenotypes. Development 125: 2273–2282 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources