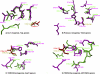Identification of functional subclasses in the DJ-1 superfamily proteins - PubMed (original) (raw)
Identification of functional subclasses in the DJ-1 superfamily proteins
Ying Wei et al. PLoS Comput Biol. 2007.
Abstract
Genomics has posed the challenge of determination of protein function from sequence and/or 3-D structure. Functional assignment from sequence relationships can be misleading, and structural similarity does not necessarily imply functional similarity. Proteins in the DJ-1 family, many of which are of unknown function, are examples of proteins with both sequence and fold similarity that span multiple functional classes. THEMATICS (theoretical microscopic titration curves), an electrostatics-based computational approach to functional site prediction, is used to sort proteins in the DJ-1 family into different functional classes. Active site residues are predicted for the eight distinct DJ-1 proteins with available 3-D structures. Placement of the predicted residues onto a structural alignment for six of these proteins reveals three distinct types of active sites. Each type overlaps only partially with the others, with only one residue in common across all six sets of predicted residues. Human DJ-1 and YajL from Escherichia coli have very similar predicted active sites and belong to the same probable functional group. Protease I, a known cysteine protease from Pyrococcus horikoshii, and PfpI/YhbO from E. coli, a hypothetical protein of unknown function, belong to a separate class. THEMATICS predicts a set of residues that is typical of a cysteine protease for Protease I; the prediction for PfpI/YhbO bears some similarity. YDR533Cp from Saccharomyces cerevisiae, of unknown function, and the known chaperone Hsp31 from E. coli constitute a third group with nearly identical predicted active sites. While the first four proteins have predicted active sites at dimer interfaces, YDR533Cp and Hsp31 both have predicted sites contained within each subunit. Although YDR533Cp and Hsp31 form different dimers with different orientations between the subunits, the predicted active sites are superimposable within the monomer structures. Thus, the three predicted functional classes form four different types of quaternary structures. The computational prediction of the functional sites for protein structures of unknown function provides valuable clues for functional classification.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
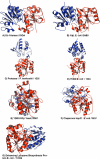
Figure 1. Dimer Structures for Seven DJ-1 Family Members with the Two Subunits Shown in Red and Blue
(A) Human DJ-1; (B) YajL from E. coli; (C) Protease I from P. horikoshii; (D) YhbO from E. coli; (E) YDR533Cp from yeast; (F) Chaperone Hsp31 from E. coli; (G) the structural genomics putative Enhancing lycopene biosynthesis protein from E. coli. For all structures, the red subunits are oriented so that they are superimposable on each other. The relative positions of the blue subunits then illustrate the different types of dimer formation.
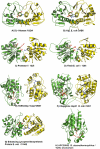
Figure 2. Ribbon Diagrams of Eight DJ-1 Family Proteins with Predicted Active Sites
(A) Human DJ-1; (B) YajL E. coli; (C) Protease I P. horikoshii; (D) YhbO E. coli; (E) YDR533Cp yeast; (F) Chaperone Hsp31 E. coli; (G) putative Enhancing lycopene biosynthesis protein E. coli; (H) APC35852 B. stearothermophilus. The subunit backbones are shown in yellow and green. Residues predicted by THEMATICS to be active site residues are shown in red (from the green subunit) and blue (from the yellow subunit).
Figure 3. Superpositions of the THEMATICS Predicted Active Site Residues (in Green and Magenta) for DJ-1 Family Members
(A) DJ-1 (magenta), YajL (green); (B) Protease I (magenta), YhbO (green); (C) Ydr533 (magenta), Hsp31 (green); (D) Ydr533 (magenta), APC35852 (green). The conserved cysteine residues that are not THEMATICS-positive residues are included for comparison purposes and are shown in yellow and red.
Similar articles
- Dissection of the dimerization modes in the DJ-1 superfamily.
Jung HJ, Kim S, Kim YJ, Kim MK, Kang SG, Lee JH, Kim W, Cha SS. Jung HJ, et al. Mol Cells. 2012 Feb;33(2):163-71. doi: 10.1007/s10059-012-2220-6. Epub 2012 Jan 2. Mol Cells. 2012. PMID: 22228183 Free PMC article. - The 1.8-A resolution crystal structure of YDR533Cp from Saccharomyces cerevisiae: a member of the DJ-1/ThiJ/PfpI superfamily.
Wilson MA, St Amour CV, Collins JL, Ringe D, Petsko GA. Wilson MA, et al. Proc Natl Acad Sci U S A. 2004 Feb 10;101(6):1531-6. doi: 10.1073/pnas.0308089100. Epub 2004 Jan 26. Proc Natl Acad Sci U S A. 2004. PMID: 14745011 Free PMC article. - Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain.
Lee SJ, Kim SJ, Kim IK, Ko J, Jeong CS, Kim GH, Park C, Kang SO, Suh PG, Lee HS, Cha SS. Lee SJ, et al. J Biol Chem. 2003 Nov 7;278(45):44552-9. doi: 10.1074/jbc.M304517200. Epub 2003 Aug 25. J Biol Chem. 2003. PMID: 12939276 - The role of cysteine oxidation in DJ-1 function and dysfunction.
Wilson MA. Wilson MA. Antioxid Redox Signal. 2011 Jul 1;15(1):111-22. doi: 10.1089/ars.2010.3481. Epub 2011 Jan 14. Antioxid Redox Signal. 2011. PMID: 20812780 Free PMC article. Review. - Characterization and prediction of protein interfaces to infer protein-protein interaction networks.
Keskin O, Tuncbag N, Gursoy A. Keskin O, et al. Curr Pharm Biotechnol. 2008 Apr;9(2):67-76. doi: 10.2174/138920108783955191. Curr Pharm Biotechnol. 2008. PMID: 18393863 Review.
Cited by
- Engineered disulfide bonds restore chaperone-like function of DJ-1 mutants linked to familial Parkinson's disease.
Logan T, Clark L, Ray SS. Logan T, et al. Biochemistry. 2010 Jul 13;49(27):5624-33. doi: 10.1021/bi902164h. Biochemistry. 2010. PMID: 20527929 Free PMC article. - Copper Metabolism in Naegleria gruberi and Its Deadly Relative Naegleria fowleri.
Ženíšková K, Grechnikova M, Sutak R. Ženíšková K, et al. Front Cell Dev Biol. 2022 Apr 11;10:853463. doi: 10.3389/fcell.2022.853463. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35478954 Free PMC article. - DJ-1 in Ocular Diseases: A Review.
Liu C, Liu X, Qi J, Pant OP, Lu CW, Hao J. Liu C, et al. Int J Med Sci. 2018 Feb 12;15(5):430-435. doi: 10.7150/ijms.23428. eCollection 2018. Int J Med Sci. 2018. PMID: 29559831 Free PMC article. Review. - Formation of a stabilized cysteine sulfinic acid is critical for the mitochondrial function of the parkinsonism protein DJ-1.
Blackinton J, Lakshminarasimhan M, Thomas KJ, Ahmad R, Greggio E, Raza AS, Cookson MR, Wilson MA. Blackinton J, et al. J Biol Chem. 2009 Mar 6;284(10):6476-85. doi: 10.1074/jbc.M806599200. Epub 2009 Jan 5. J Biol Chem. 2009. PMID: 19124468 Free PMC article. - Protein aggregation in a mutant deficient in yajL, the bacterial homolog of the Parkinsonism-associated protein DJ-1.
Kthiri F, Le HT, Gautier V, Caldas T, Malki A, Landoulsi A, Bohn C, Bouloc P, Richarme G. Kthiri F, et al. J Biol Chem. 2010 Apr 2;285(14):10328-36. doi: 10.1074/jbc.M109.077529. Epub 2010 Feb 2. J Biol Chem. 2010. PMID: 20124404 Free PMC article.
References
- Ko J, Murga LF, Andre P, Yang H, Ondrechen MJ, et al. Statistical criteria for the identification of protein active sites using theoretical microscopic titration curves. Proteins. 2005;59:183–195. - PubMed
- Ko J, Murga LF, We Yi, Ondrechen MJ. Prediction of active sites for protein structures from computed chemical properties. Bioinformatics. 2005;21:i258–265. - PubMed
- Murga LF, Wei Y, Andre P, Clifton JG, Ringe D, et al. Physicochemical methods for prediction of functional information for proteins. Israel J Chem. 2004;44:299–308.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous