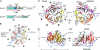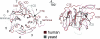Crystal structure of the N-terminal domain of the human protooncogene Nup214/CAN - PubMed (original) (raw)
Comparative Study
. 2007 Feb 6;104(6):1783-8.
doi: 10.1073/pnas.0610828104. Epub 2007 Jan 30.
Affiliations
- PMID: 17264208
- PMCID: PMC1794303
- DOI: 10.1073/pnas.0610828104
Comparative Study
Crystal structure of the N-terminal domain of the human protooncogene Nup214/CAN
Johanna Napetschnig et al. Proc Natl Acad Sci U S A. 2007.
Abstract
The mammalian nuclear pore complex (NPC) is an approximately 120-MDa proteinaceous assembly consisting of approximately 30 proteins and is the sole gate in the nuclear envelope. The human protooncogene Nup214 was first identified as a target for chromosomal translocation involved in leukemogenesis. Nup214 is located on the cytoplasmic face of the NPC and is implicated in anchoring the cytoplasmic filaments of the NPC and recruiting the RNA helicase Ddx19. Here, we present the crystal structure of the human Nup214 N-terminal domain at 1.65-A resolution. The structure reveals a seven-bladed beta-propeller followed by a 30-residue C-terminal extended peptide segment, which folds back onto the beta-propeller and binds to its bottom face. The beta-propeller repeats lack any recognizable sequence motif and are distinguished by extensive insertions between the canonical beta-strands. We propose a mechanism by which the C-terminal peptide extension is involved in NPC assembly.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Fig. 1.
The structure of the NTD of human Nup214. (A) Domain structure of Nup214 and Nup159. The construct used for crystallization is boxed red, and two phosphorylation sites of the NTD are indicated. Residues observed in the crystal structures are boxed in blue. (B) Schematic representation of the NTD structure. The blades of the β-propeller are labeled from 1 to 7. The CTE is shown in blue, and β-strands forming the double-Velcro closure are indicated with an asterisk. (C) Ribbon representation of the NTD structure. A 180°-rotated view is shown on the right. As a reference, the strands of blade 3 are labeled A–D. The blades of the β-propeller and the CTE are labeled as in B. The helical insertions are shown in pink. (D) Ribbon representation of side views of the structure of the NTD. The view on the right is rotated by 180°.
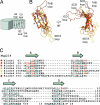
Fig. 2.
Superposition of the NTD β-propeller blades. (A) Schematic drawing of a β-propeller fold indicating the β-strands and loops of one β-propeller blade. (B) Coil representation of the structural alignment of the seven blades of the β-propeller. Blades are colored as in Fig. 1. As a reference, the Cα atoms of blade 2 are shown as orange spheres. A 90°-rotated view is shown on the right. (C) Structure-based sequence alignment of the blades. The β-strands are indicated above the sequence. Similar residues are shown in red, and the residues of each blade that participate in β-sheet hydrogen bonds are underlined in gray.
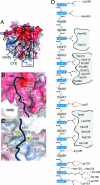
Fig. 3.
CTE binding to the bottom face of the β-propeller. (A) The surface of the Nup214 β-propeller is colored according to the electrostatic potential from −10 kBT (red) to + 10 kBT (blue). The CTE is shown in blue coil representation with the side chains in ball-and-stick representation. The black box indicates the region magnified in D. (B) Hydrophobic interactions of CTE residues Val-410, Leu-413, and Leu-414 (yellow). (C) Interactions of Leu-420 and Leu-422 (yellow) with residues of the β-propeller. Hydrophobic pocket-forming residues are shown in gray. The surface of the β-propeller is colored as in A. (D) Schematic representation of the contacts between the β-propeller and the CTE. Hydrogen and ionic bonds are indicated by orange dashed lines and van der Waals contacts with gray grooves.
Fig. 4.
Structural comparison of the NTD of the human Nup214 and its yeast homolog Nup159. Cα trace of a structural superposition of the Nup214 NTD (ruby) and the Nup159 β-propeller (gray).
Fig. 5.
Conserved features of the NTD. (A) Surface representation showing conservation of residues within higher eukaryotes. The conserved surface is shaded from gray (<70% identity) to red (100% identity) according to the alignment in
SI Fig. 8
. (B) Electrostatic potential of the NTD surface (colored as in Fig. 3_A_). (C) Surface representation of the bottom face of the Nup2141–405 showing the surface conservation within higher eukaryotes (Left) and electrostatic potential (Right). The Cα trace of the CTE is shown in a blue coil representation.
Similar articles
- Molecular basis for the anchoring of proto-oncoprotein Nup98 to the cytoplasmic face of the nuclear pore complex.
Stuwe T, von Borzyskowski LS, Davenport AM, Hoelz A. Stuwe T, et al. J Mol Biol. 2012 Jun 22;419(5):330-46. doi: 10.1016/j.jmb.2012.03.024. Epub 2012 Apr 2. J Mol Biol. 2012. PMID: 22480613 Free PMC article. - Structural evolution of the membrane-coating module of the nuclear pore complex.
Liu X, Mitchell JM, Wozniak RW, Blobel G, Fan J. Liu X, et al. Proc Natl Acad Sci U S A. 2012 Oct 9;109(41):16498-503. doi: 10.1073/pnas.1214557109. Epub 2012 Sep 26. Proc Natl Acad Sci U S A. 2012. PMID: 23019579 Free PMC article. - The N-terminal domain of Nup159 forms a beta-propeller that functions in mRNA export by tethering the helicase Dbp5 to the nuclear pore.
Weirich CS, Erzberger JP, Berger JM, Weis K. Weirich CS, et al. Mol Cell. 2004 Dec 3;16(5):749-60. doi: 10.1016/j.molcel.2004.10.032. Mol Cell. 2004. PMID: 15574330 - Membrane-coating lattice scaffolds in the nuclear pore and vesicle coats: commonalities, differences, challenges.
Leksa NC, Schwartz TU. Leksa NC, et al. Nucleus. 2010 Jul-Aug;1(4):314-8. doi: 10.4161/nucl.1.4.11798. Epub 2010 Mar 12. Nucleus. 2010. PMID: 21327078 Free PMC article. Review. - Toward the atomic structure of the nuclear pore complex: when top down meets bottom up.
Hoelz A, Glavy JS, Beck M. Hoelz A, et al. Nat Struct Mol Biol. 2016 Jul;23(7):624-30. doi: 10.1038/nsmb.3244. Epub 2016 Jun 6. Nat Struct Mol Biol. 2016. PMID: 27273515 Free PMC article. Review.
Cited by
- Structural and functional analysis of Nup120 suggests ring formation of the Nup84 complex.
Seo HS, Ma Y, Debler EW, Wacker D, Kutik S, Blobel G, Hoelz A. Seo HS, et al. Proc Natl Acad Sci U S A. 2009 Aug 25;106(34):14281-6. doi: 10.1073/pnas.0907453106. Epub 2009 Aug 11. Proc Natl Acad Sci U S A. 2009. PMID: 19706512 Free PMC article. - The nucleoporin Gle1 activates DEAD-box protein 5 (Dbp5) by promoting ATP binding and accelerating rate limiting phosphate release.
Gray S, Cao W, Montpetit B, De La Cruz EM. Gray S, et al. Nucleic Acids Res. 2022 Apr 22;50(7):3998-4011. doi: 10.1093/nar/gkac164. Nucleic Acids Res. 2022. PMID: 35286399 Free PMC article. - Insights into the role of Nup62 and Nup93 in assembling cytoplasmic ring and central transport channel of the nuclear pore complex.
Madheshiya PK, Shukla E, Singh J, Bawaria S, Ansari MY, Chauhan R. Madheshiya PK, et al. Mol Biol Cell. 2022 Dec 1;33(14):ar139. doi: 10.1091/mbc.E22-01-0027. Epub 2022 Oct 12. Mol Biol Cell. 2022. PMID: 36222862 Free PMC article. - Functionalization of a nanopore: the nuclear pore complex paradigm.
Peters R. Peters R. Biochim Biophys Acta. 2009 Oct;1793(10):1533-9. doi: 10.1016/j.bbamcr.2009.06.003. Epub 2009 Jul 9. Biochim Biophys Acta. 2009. PMID: 19596381 Free PMC article. Review. - Molecular basis for the anchoring of proto-oncoprotein Nup98 to the cytoplasmic face of the nuclear pore complex.
Stuwe T, von Borzyskowski LS, Davenport AM, Hoelz A. Stuwe T, et al. J Mol Biol. 2012 Jun 22;419(5):330-46. doi: 10.1016/j.jmb.2012.03.024. Epub 2012 Apr 2. J Mol Biol. 2012. PMID: 22480613 Free PMC article.
References
- Stoffler D, Feja B, Fahrenkrog B, Walz J, Typke D, Aebi U. J Mol Biol. 2003;328:119–130. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases