Evolutionary insights into the ecology of coronaviruses - PubMed (original) (raw)
Evolutionary insights into the ecology of coronaviruses
D Vijaykrishna et al. J Virol. 2007 Apr.
Erratum in
- J Virol. 2007 Aug;81(15):8371
Abstract
Although many novel members of the Coronaviridae have recently been recognized in different species, the ecology of coronaviruses has not been established. Our study indicates that bats harbor a much wider diversity of coronaviruses than any other animal species. Dating of different coronavirus lineages suggests that bat coronaviruses are older than those recognized in other animals and that the human severe acute respiratory syndrome (SARS) coronavirus was directly derived from viruses from wild animals in wet markets of southern China. Furthermore, the most closely related bat and SARS coronaviruses diverged in 1986, an estimated divergence time of 17 years prior to the outbreak, suggesting that there may have been transmission via an unknown intermediate host. Analysis of lineage-specific selection pressure also indicated that only SARS coronaviruses in civets and humans were under significant positive selection, also demonstrating a recent interspecies transmission. Analysis of population dynamics revealed that coronavirus populations in bats have constant population growth, while viruses from all other hosts show epidemic-like increases in population. These results indicate that diverse coronaviruses are endemic in different bat species, with repeated introductions to other animals and occasional establishment in other species. Our findings suggest that bats are likely the natural hosts for all presently known coronavirus lineages and that all coronaviruses recognized in other species were derived from viruses residing in bats. Further surveillance of bat and other animal populations is needed to fully describe the ecology and evolution of this virus family.
Figures
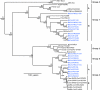
FIG. 1.
Phylogenetic relationships of helicase (A), spike (B), and nucleocapsid (C) genes based on alignments of 1,797, 2,204, and 1,812 nucleotides, respectively. Breda virus (AY427798) was used to root the trees. Numbers at branch nodes indicate neighbor-joining bootstrap values of ≥50%. Nodes with Bayesian posterior probabilities of ≤95% are indicated with asterisks. Bar, 0.1 nucleotide substitution per site (A) or 100 nucleotide substitutions per site (B and C).
FIG. 2.
Divergence dates of coronavirus lineages represented on a phylogenetic tree generated based on 1,797 nucleotides of the complete HEL domain sequence. Breda virus (AY427798) was used to root the tree. Numbers at branch nodes indicate the times of divergence, calculated using the uncorrelated exponential relaxed-clock model in BEAST v1.4. For an explanation of the letters at branch nodes, see Table 2.
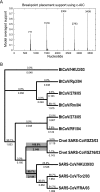
FIG. 3.
Detection of recombination (A) and lineage-specific selection pressure (B) on the spike genes of SARS-CoVs. Mean ω values calculated using GA are presented above branches, with ω values of >1 shown in bold. Numbers below branches indicate an averaged model probability of >1 along specific lineages. Branches with P values of ≥95% are highlighted in gray. The Akaike information criterion (c-AIC) for the best-fitting model was 23,557.
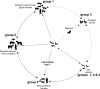
FIG. 4.
Postulated ecology of coronaviruses. Solid and dashed lines represent confirmed and hypothetical interspecies transmission pathways, respectively, and arrows represent evolutionary pathways.
Similar articles
- Bat origin of human coronaviruses.
Hu B, Ge X, Wang LF, Shi Z. Hu B, et al. Virol J. 2015 Dec 22;12:221. doi: 10.1186/s12985-015-0422-1. Virol J. 2015. PMID: 26689940 Free PMC article. Review. - Identification of diverse alphacoronaviruses and genomic characterization of a novel severe acute respiratory syndrome-like coronavirus from bats in China.
He B, Zhang Y, Xu L, Yang W, Yang F, Feng Y, Xia L, Zhou J, Zhen W, Feng Y, Guo H, Zhang H, Tu C. He B, et al. J Virol. 2014 Jun;88(12):7070-82. doi: 10.1128/JVI.00631-14. Epub 2014 Apr 9. J Virol. 2014. PMID: 24719429 Free PMC article. - Discovery of seven novel Mammalian and avian coronaviruses in the genus deltacoronavirus supports bat coronaviruses as the gene source of alphacoronavirus and betacoronavirus and avian coronaviruses as the gene source of gammacoronavirus and deltacoronavirus.
Woo PC, Lau SK, Lam CS, Lau CC, Tsang AK, Lau JH, Bai R, Teng JL, Tsang CC, Wang M, Zheng BJ, Chan KH, Yuen KY. Woo PC, et al. J Virol. 2012 Apr;86(7):3995-4008. doi: 10.1128/JVI.06540-11. Epub 2012 Jan 25. J Virol. 2012. PMID: 22278237 Free PMC article. - Presence of Recombinant Bat Coronavirus GCCDC1 in Cambodian Bats.
Zhu F, Duong V, Lim XF, Hul V, Chawla T, Keatts L, Goldstein T, Hassanin A, Tu VT, Buchy P, Sessions OM, Wang LF, Dussart P, Anderson DE. Zhu F, et al. Viruses. 2022 Jan 18;14(2):176. doi: 10.3390/v14020176. Viruses. 2022. PMID: 35215769 Free PMC article. - Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS.
Drexler JF, Corman VM, Drosten C. Drexler JF, et al. Antiviral Res. 2014 Jan;101:45-56. doi: 10.1016/j.antiviral.2013.10.013. Epub 2013 Oct 31. Antiviral Res. 2014. PMID: 24184128 Free PMC article. Review.
Cited by
- Genetic and Phylogenetic Analysis of Feline Coronavirus in Guangxi Province of China from 2021 to 2024.
Shi K, He M, Shi Y, Long F, Shi Y, Yin Y, Pan Y, Li Z, Feng S. Shi K, et al. Vet Sci. 2024 Sep 25;11(10):455. doi: 10.3390/vetsci11100455. Vet Sci. 2024. PMID: 39453047 Free PMC article. - Recent evolutionary origin and localized diversity hotspots of mammalian coronaviruses.
Maestri R, Perez-Lamarque B, Zhukova A, Morlon H. Maestri R, et al. Elife. 2024 Aug 28;13:RP91745. doi: 10.7554/eLife.91745. Elife. 2024. PMID: 39196812 Free PMC article. - SARS-CoV and SARS-CoV -2 cross-reactive antibodies in domestic animals and wildlife in Nigeria suggest circulation of sarbecoviruses.
Agusi ER, Schön J, Allendorf V, Eze EA, Asala O, Shittu I, Balkema-Buschmann A, Wernike K, Tekki I, Ofua M, Adefegha O, Olubade O, Ogunmolawa O, Dietze K, Globig A, Hoffmann D, Meseko CA. Agusi ER, et al. One Health. 2024 Mar 12;18:100709. doi: 10.1016/j.onehlt.2024.100709. eCollection 2024 Jun. One Health. 2024. PMID: 38533194 Free PMC article. - Comparative genomics of spike, envelope, and nucleocapsid protein of severe acute respiratory syndrome coronavirus 2.
Khan SS, Ullah A. Khan SS, et al. Afr Health Sci. 2023 Sep;23(3):384-399. doi: 10.4314/ahs.v23i3.45. Afr Health Sci. 2023. PMID: 38357143 Free PMC article. - Recombination in sarbecovirus lineage and mutations/insertions in spike protein are linked to the emergence and adaptation of SARS-CoV-2.
Som A, Sharma AK, Kumari P. Som A, et al. Bioinformation. 2022 Oct 31;18(10):951-961. doi: 10.6026/97320630018951. eCollection 2022. Bioinformation. 2022. PMID: 37693920 Free PMC article.
References
- Arbogast, B. S., S. V. Edwards, J. Wakeley, P. Beerli, and J. B. Slowinski. 2002. Estimating divergence times from molecular data on phylogenetic and population genetic timescales. Annu. Rev. Ecol. Syst. 33:707-740.
- Avise, J. C., J. Arnold, R. M. Ball, E. Bermingham, T. Lamb, J. E. Neigel, C. A. Reeb, and N. C. Saunders. 1987. Intraspecific phylogeography: the mitochondrial DNA bridge between population genetics and systematics. Annu. Rev. Ecol. Syst. 18:489-522.
- Baker, J. E. 1962. The manner and efficiency of raptor depredations on bats. Condor 64:500-504.
- Chua, K. B., W. J. Bellini, P. A. Rota, B. H. Harcourt, A. Tamin, S. K. Lam, T. G. Ksiazej, P. E. Rollin, S. R. Zaki, W. J. Shieh, C. S. Goldsmith, D. J. Gubler, J. T. Roehrig, B. Eaton, A. R. Gould, J. Olson, H. Field, P. Daniels, A. E. Ling, C. J. Peters, L. J. Anderson, and B. W. J. Mahy. 2000. Nipah virus: a recently emergent deadly paramyxovirus. Science 288:1432-1435. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous