Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation - PubMed (original) (raw)
doi: 10.1086/512203. Epub 2007 Jan 29.
Heinrich Sticht, Gerhard Hammersen, Gabriele Gillessen-Kaesbach, David R Fitzpatrick, Gudrun Nürnberg, Frank Brasch, Heidemarie Schirmer-Zimmermann, John L Tolmie, David Chitayat, Gunnar Houge, Lorena Fernández-Martínez, Sarah Keating, Geert Mortier, Raoul C M Hennekam, Axel von der Wense, Anne Slavotinek, Peter Meinecke, Pierre Bitoun, Christian Becker, Peter Nürnberg, André Reis, Anita Rauch
Affiliations
- PMID: 17273977
- PMCID: PMC1821097
- DOI: 10.1086/512203
Mutations in STRA6 cause a broad spectrum of malformations including anophthalmia, congenital heart defects, diaphragmatic hernia, alveolar capillary dysplasia, lung hypoplasia, and mental retardation
Francesca Pasutto et al. Am J Hum Genet. 2007 Mar.
Abstract
We observed two unrelated consanguineous families with malformation syndromes sharing anophthalmia and distinct eyebrows as common signs, but differing for alveolar capillary dysplasia or complex congenital heart defect in one and diaphragmatic hernia in the other family. Homozygosity mapping revealed linkage to a common locus on chromosome 15, and pathogenic homozygous mutations were identified in STRA6, a member of a large group of "stimulated by retinoic acid" genes encoding novel transmembrane proteins, transcription factors, and secreted signaling molecules or proteins of largely unknown function. Subsequently, homozygous STRA6 mutations were also demonstrated in 3 of 13 patients chosen on the basis of significant phenotypic overlap to the original cases. While a homozygous deletion generating a premature stop codon (p.G50AfsX22) led to absence of the immunoreactive protein in patient's fibroblast culture, structural analysis of three missense mutations (P90L, P293L, and T321P) suggested significant effects on the geometry of the loops connecting the transmembrane helices of STRA6. Two further variations in the C-terminus (T644M and R655C) alter specific functional sites, an SH2-binding motif and a phosphorylation site, respectively. STRA6 mutations thus define a pleiotropic malformation syndrome representing the first human phenotype associated with mutations in a gene from the "STRA" group.
Figures
Figure 1.
A and B, Pedigrees of the two families and segregation of the respective mutation (M). Arrows indicate the probands. Individuals marked with asterisks participated in the genomewide linkage analysis. C and D, Examples of electropherograms of STRA6 mutations in families 1 (C) and 2 (D). WT = Wild type.
Figure 2.
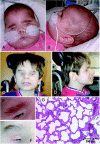
A–D, Frontal and lateral views of patient IV:2 of family 1 at age 6 mo (A and B) and patient IV:1 of family 2 at age 13 years (C and D). Note similar mild dysmorphism with broad, flaring, and only upward-growing eyebrows; broad nasal bridge; large, low-set ears; and receding chin. E and F, Close-up of right eyebrows of IV:2 of family 1 (E) and IV:1 of family 2 (F). G, Hematoxylin-eosin staining of lung biopsy, showing deficiency in the number of alveolar units and pulmonary capillary vessels with thickening of the interalveolar septa.
Figure 3.
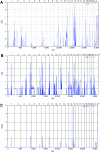
Graphic summaries of multipoint linkage analyses in families 1 and 2, performed using the Allegro program under the assumption of autosomal recessive inheritance with full penetrance. A, Genomewide multipoint LOD scores of family 1, including genotypes from individuals III:1–4, IV:1–3, and IV:5–6 (fig. 1_A_), identifying a single locus on chromosome 15 with the expected maximum LOD score of 2.9. B, Genomewide multipoint LOD scores of family 2, including genotypes from individuals III:1–2 and IV:1–3 (fig. 1_B_), identifying multiple loci on chromosomes 1, 4, 7, 8, and 15 with the expected maximum LOD score of 1.9. C, Summary LOD scores including both families allowing also for locus heterogeneity (HLOD) revealing a single locus on chromosome 15 with a maximum LOD and HLOD of 4.8.
Figure 4.
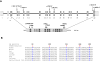
Haplotype analyses on chromosome 15q23-25.1, showing homozygous markers linked to the disease locus in family 1 (A) and family 2 (B). According to the NCBI human genome overview page, build 35.1, the flanking markers SNP_A-1511966 (rs1822829) and SNP_A-1509050 (rs1077965) span a region of ∼12 Mb between 65.21 and 77.85 Mb from pter.
Figure 5.
Location and conservation of mutated residues. A, Location of mutations along STRA6 genomic and protein schematic representation, respectively. Transmembrane helices predicted by all three algorithms used are shown as dark gray boxes; helices predicted only by one or two of the algorithms are shown as dotted light gray boxes. B, ClustalW multiple sequence alignments of human STRA6 regions to orthologues encompassing mutated residues. All these residues are evolutionary conserved. The respective substituted amino acid is highlighted in blue.
Figure 6.
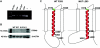
Characterization of mutations. A, STRA6 RT-PCR on cultured fibroblast cells from the affected fetus (IV:3, family 2) and from a healthy individual (C) grown in the absence and the presence of puromycin (p−/p+) as inhibitor of translation, respectively, showing no evidence for early nonsense-mediated mRNA decay; (+) positive control, (−) negative control. B, Western blot analysis of cultured fibroblast protein cell extract showing a STRA6 protein band in two human healthy control lanes (indicated as “WT”) but not in the two lanes with the homozygous mutant c.145_147delC (from IV:3, family 2). C, Model showing the effect of the P293L mutation (right) on the STRA6 structure in comparison with the wild type (left). The membrane is indicated by a dotted line. The transmembrane helix (A300-V319) that is proximal to the site of the mutation is shown in ribbon presentation, and the two adjacent helices are indicated by cylinders. In the wild type, residue P293 represents the N-terminal cap of a helix starting at L294, whereas the L293 present in the mutant allows an N-terminal extension of the helix, now starting at H291 and changing the topology of the respective loop. Therefore, the P293L mutation is predicted to cause an extension of the helix by three residues, thus affecting the structure and orientation of the respective loop. Moreover, it is important to note that V319 is also the N-terminal residue of an extremely short loop (maximum predicted length between aa 319 and aa 326), in which a second mutation (T321P) has been identified.
Figure 7.
Expression pattern of STRA6 determined by RT-PCR in normal adult human (A) and eye (B) tissues. RPE = Retinal pigment epithelium; TM = Trabecular meshwork.
Similar articles
- Pulmonary hypoplasia-diaphragmatic hernia-anophthalmia-cardiac defect (PDAC) syndrome due to STRA6 mutations--what are the minimal criteria?
Segel R, Levy-Lahad E, Pasutto F, Picard E, Rauch A, Alterescu G, Schimmel MS. Segel R, et al. Am J Med Genet A. 2009 Nov;149A(11):2457-63. doi: 10.1002/ajmg.a.33038. Am J Med Genet A. 2009. PMID: 19839040 - A novel mutation in two Hmong families broadens the range of STRA6-related malformations to include contractures and camptodactyly.
Marcadier JL, Mears AJ, Woods EA, Fisher J, Airheart C, Qin W, Beaulieu CL, Dyment DA, Innes AM, Curry CJ; Care4Rare Canada Consortium. Marcadier JL, et al. Am J Med Genet A. 2016 Jan;170A(1):11-8. doi: 10.1002/ajmg.a.37389. Epub 2015 Sep 16. Am J Med Genet A. 2016. PMID: 26373900 - Matthew-Wood syndrome is caused by truncating mutations in the retinol-binding protein receptor gene STRA6.
Golzio C, Martinovic-Bouriel J, Thomas S, Mougou-Zrelli S, Grattagliano-Bessieres B, Bonniere M, Delahaye S, Munnich A, Encha-Razavi F, Lyonnet S, Vekemans M, Attie-Bitach T, Etchevers HC. Golzio C, et al. Am J Hum Genet. 2007 Jun;80(6):1179-87. doi: 10.1086/518177. Epub 2007 Apr 11. Am J Hum Genet. 2007. PMID: 17503335 Free PMC article. - TMCO1 deficiency causes autosomal recessive cerebrofaciothoracic dysplasia.
Alanay Y, Ergüner B, Utine E, Haçariz O, Kiper PO, Taşkıran EZ, Perçin F, Uz E, Sağiroğlu MŞ, Yuksel B, Boduroglu K, Akarsu NA. Alanay Y, et al. Am J Med Genet A. 2014 Feb;164A(2):291-304. doi: 10.1002/ajmg.a.36248. Epub 2013 Nov 5. Am J Med Genet A. 2014. PMID: 24194475 Review. - Autosomal recessive congenital diaphragmatic defects in the Arabs.
Farag TI, Bastaki L, Marafie M, al-Awadi SA, Krsz J. Farag TI, et al. Am J Med Genet. 1994 Apr 15;50(3):300-1. doi: 10.1002/ajmg.1320500316. Am J Med Genet. 1994. PMID: 8042677 Review. No abstract available.
Cited by
- Dynamic upregulation of retinoic acid signal in the early postnatal murine heart promotes cardiomyocyte cell cycle exit and maturation.
Fujikawa Y, Kato K, Unno K, Narita S, Okuno Y, Sato Y, Takefuji M, Murohara T. Fujikawa Y, et al. Sci Rep. 2024 Aug 30;14(1):20222. doi: 10.1038/s41598-024-70918-1. Sci Rep. 2024. PMID: 39215116 Free PMC article. - The Logistical Backbone of Photoreceptor Cell Function: Complementary Mechanisms of Dietary Vitamin A Receptors and Rhodopsin Transporters.
Leung M, Steinman J, Li D, Lor A, Gruesen A, Sadah A, van Kuijk FJ, Montezuma SR, Kondkar AA, Radhakrishnan R, Lobo GP. Leung M, et al. Int J Mol Sci. 2024 Apr 12;25(8):4278. doi: 10.3390/ijms25084278. Int J Mol Sci. 2024. PMID: 38673863 Free PMC article. Review. - Vitamin A deficiency compromises the barrier function of the retinal pigment epithelium.
Moon J, Zhou G, Jankowsky E, von Lintig J. Moon J, et al. PNAS Nexus. 2023 May 19;2(6):pgad167. doi: 10.1093/pnasnexus/pgad167. eCollection 2023 Jun. PNAS Nexus. 2023. PMID: 37275262 Free PMC article. - Mapping of the extracellular RBP4 ligand binding domain on the RBPR2 receptor for Vitamin A transport.
Radhakrishnan R, Leung M, Solanki AK, Lobo GP. Radhakrishnan R, et al. Front Cell Dev Biol. 2023 Feb 22;11:1105657. doi: 10.3389/fcell.2023.1105657. eCollection 2023. Front Cell Dev Biol. 2023. PMID: 36910150 Free PMC article. - Missense Mutations in MAB21L1: Causation of Novel Autosomal Dominant Ocular BAMD Syndrome.
Wang P, Wu P, Wang J, Zeng Y, Jiang Y, Wang Y, Li S, Xiao X, Zhang Q. Wang P, et al. Invest Ophthalmol Vis Sci. 2023 Mar 1;64(3):19. doi: 10.1167/iovs.64.3.19. Invest Ophthalmol Vis Sci. 2023. PMID: 36892533 Free PMC article. Review.
References
Web Resources
- ClustalW, http://www.ebi.ac.uk/clustalw/
- ELM, The Eukaryotic Linear Motif Resource for Functional Sites in Proteins, http://elm.eu.org/
- SwissProt, http://ca.expasy.org/sprot/ (for reference sequence Q8TB21_Human)
- National Center for Biotechnology Information (NCBI), http://www.ncbi.nlm.nih.gov/
- NPS@ Consensus Secondary Structure Prediction, http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_seccons...
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases