Cdc7-Dbf4 and the human S checkpoint response to UVC - PubMed (original) (raw)
Cdc7-Dbf4 and the human S checkpoint response to UVC
Timothy P Heffernan et al. J Biol Chem. 2007.
Abstract
The S checkpoint response to ultraviolet radiation (UVC) that inhibits replicon initiation is dependent on the ATR and Chk1 kinases. Downstream effectors of this response, however, are not well characterized. Data reported here eliminated Cdc25A degradation and inhibition of Cdk2-cyclin E as intrinsic components of the UVC-induced pathway of inhibition of replicon initiation in human cells. A sublethal dose of UVC (1 J/m(2)), which selectively inhibits replicon initiation by 50%, failed to reduce the amount of Cdc25A protein or decrease Cdk2-cyclin E kinase activity. Cdc25A degradation was observed after irradiation with cytotoxic fluences of UVC, suggesting that severe inhibition of DNA chain elongation and activation of the replication checkpoint might be responsible for the UVC-induced degradation of Cdc25A. Another proposed effector of the S checkpoint is the Cdc7-Dbf4 complex. Dbf4 interacted weakly with Chk1 in vivo but was recognized as a substrate for Chk1-dependent phosphorylation in vitro. FLAG-Dbf4 formed complexes with endogenous Cdc7, and this interaction was stable in UVC-irradiated HeLa cells. Overexpression of FLAG- or Myc-tagged Dbf4 abrogated the S checkpoint response to UVC but not ionizing radiation. These findings implicate a Dbf4-dependent kinase as a possible target of the ATR- and Chk1-dependent S checkpoint response to UVC.
Figures
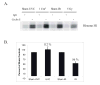
Fig. 1. UVC and IR activate different signaling intermediates in human cells
A.) NHF1 cells were grown in the presence of [14C]thymidine for ~ 40 h to label DNA uniformly, and then in non-radioactive medium overnight. Cells were sham-treated or exposed to UVC (1 J/m2) or IR (5 Gy), incubated at 37 °C for 30 min, and then labeled for 15 min in medium containing [3H]thymidine. Cells were harvested and nascent DNA separated by velocity sedimentation. Net 3H radioactivity corrected for 14C spillover was normalized to cell number (total 14C radioactivity). Closed circles (●) represent profiles from sham-treated cells while grey circles (○) represent those from irradiated cultures. B.) Normal human fibroblasts were sham treated or irradiated with either 1 J/m2 UVC or 5 Gy IR. Cells were harvested 1 h after irradiation and cell extracts prepared for western immunoblot analysis.
Fig. 2. Cdc25A degradation is not required for the UVC-induced S checkpoint response
A.) NHF1 and HeLa cells were sham treated or irradiated with either 1 J/m2 UVC or 5 Gy IR. Cells were harvested 1 h after irradiation and cell extracts prepared for western immunoblot analysis. B.) NHF1 cells were pretreated with either DMSO or 25 μg/ml LLnL for 30 min. Cells were then sham treated or irradiated with either 1 J/m2 UVC or 5 Gy IR. Cells were harvested 1 h after irradiation and cell extracts prepared for western immunoblot analysis. C.) NHF1 cells were grown in the presence of [14C]thymidine to label DNA uniformly, then in non-radioactive medium overnight, and pretreated with DMSO or LLnL (as indicated in B). After sham treatment or irradiation with either 1 J/m2 UVC or 5 Gy IR, cells were incubated at 37 °C for 30 min, and then labeled for 15 min in medium containing [3H]thymidine. DNA synthesis activity was measured by the ratios of 3H/14C radioactivity in acid-precipitable material from cell lysates. Net 3H radioactivity was corrected for 14C spillover before normalized to cell number (total 14C radioactivity). The 3H/14C values were expressed as percentages of the paired, sham-treated controls (n=3; black bars, sham-treated controls; white bars, cell exposed to IR or UVC, as indicated; error bars correspond to one standard deviation of the mean.).
Fig. 3. Cdk2/cyclinE inhibition is not associated with activation of the UVC-induced S checkpoint
A.) NHF1 cells were sham treated or irradiated with either 1 J/m2 UVC or 5 Gy IR. Cells were harvested 1 h after irradiation and cell extracts prepared. Cdk2/cyclin E complexes were immunoprecipitated and kinase activity measured in vitro against histone H1. In control reactions, the same extracts were incubated with non-specific IgG. B.) Compilation data of 4 independent experiments were graphed as percent of sham-treated controls. Error bars represent one standard deviation of the mean.
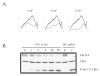
Fig. 4. UVC-induced Cdc25A degradation is a high-dose effect
A.) NHF1 cells were treated with 0, 1, or 8 J/m2 UVC and inhibition of DNA synthesis determined by velocity sedimentation as described in the legend to Fig. 1 (27). Closed diamonds (♦) represent sham treated cultures, while grey circles (○) represent UVC-irradiated cultures. B.) NHF1 cells were sham treated or irradiated with increasing fluences of UVC (0–50 J/m2) and harvested 1 h later. Parallel cultures were incubated for 24 h in medium containing 2 mM HU. Cells were harvested and extracts prepared for western immunoblot analysis.
Fig. 5. Chk1 phosphorylates Dbf4 in vitro and interacts with Dbf4 in vivo
A.) HEK-293T cells were transfected with either Flag-Chk1WT, Flag-Chk1KD, or Flag-Dbf4 expression vectors. Forty-eight hours after transfection, cells were harvested and Flag-tagged proteins immuno-precipitated and eluted as described. Flag-Dbf4 was incubated with Flag-Chk1WT or Flag-Chk1KD, for 30 min in the presence of [32P]ATP. Control reactions included only one of these proteins. The incubation was terminated upon addition of Laemmli sample buffer and proteins separated by SDS-PAGE. The gel was dried and exposed to a phosphoscreen. B.) HEK-293T cells were transfected with the indicated cDNA’s. Forty eight hours after transfection, cells were harvested and extracts were prepared. Flag-tagged proteins were immuno-precipitated with anti-Flag agarose and eluted with 200 μg/ml Flag peptide. Proteins were separated by SDS-PAGE and immuno-blotted with antibodies against the indicated proteins.
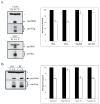
Fig. 6. Over-expression of Dbf4 reverses the UVC-induced S checkpoint
A.) HeLa cells were mock-transfected (M), transfected with an empty vector (V), or transfected with Flag-, or Myc-tagged Dbf4 expression vectors (F4). Cells were harvested 48 h later and extracts prepared for western immunoblot analysis (* = non-specific band). In separate experiments, transfected cultures were incubated with [14C]thymidine to label DNA uniformly; then, cells were treated with either 0 or 1 J/m2 UVC, incubated for 30 min at 37°C and pulsed-labeled with [3H]thymidine for 15 min. DNA synthesis was measured as described in the legend to Fig. 2C and graphed as percentages of paired, sham-treated controls [n=5 (Mock), n=7 (Vector), n=5 (Flag), n=2 (Myc); black bar, sham-treated controls; white bar, UVC-irradiated cells. Error bars correspond to one standard deviation of the mean]. B.) HeLa cells were transiently transfected with either empty vector (V) or a Flag-Dbf4 expressing vector (F4). Cells were harvested 48 h later and extracts prepared for western immunoblot analysis (* = non-specific band). In separate experiments, transfected cultures were incubated with [14C]thymidine to label DNA uniformly; then, cells were sham-treated or irradiated with either 1 J/m2 UVC or 5 Gy IR, incubated for 30 min at 37°C and pulse-labeled with [3H]thymidine for 15 min. DNA synthesis was measured as described in the legend to Fig. 2C and graphed as a percentages of paired, sham-treated controls (n=3; black bar, sham-treated controls; white bar, UVC- or IR-treated cells. Error bars correspond to one standard deviation of the mean).
Fig. 7. Flag-Dbf4 forms a complex with endogenous Cdc7 following DNA damage
HeLa cells were transiently transfected with a Flag-Dbf4 expression vector. Forty-eight hours later, cells were treated with 8 J/m2 UVC, 10 μM etoposide, or 2 mM HU. Cells were harvested 1 h later and Flag-Dbf4 immuno-precipitated with anti-Flag agarose (NS corresponds to immuno-precipitation with normal mouse IgG). The effect of DNA damage on Cdc7/Dbf4 complex formation was measured by western immunoblot analysis.
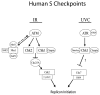
Fig. 8
Schematic representation of S checkpoint signaling in human cells.
Similar articles
- An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage.
Heffernan TP, Simpson DA, Frank AR, Heinloth AN, Paules RS, Cordeiro-Stone M, Kaufmann WK. Heffernan TP, et al. Mol Cell Biol. 2002 Dec;22(24):8552-61. doi: 10.1128/MCB.22.24.8552-8561.2002. Mol Cell Biol. 2002. PMID: 12446774 Free PMC article. - Checkpoint regulation of replication dynamics in UV-irradiated human cells.
Chastain PD 2nd, Heffernan TP, Nevis KR, Lin L, Kaufmann WK, Kaufman DG, Cordeiro-Stone M. Chastain PD 2nd, et al. Cell Cycle. 2006 Sep;5(18):2160-7. doi: 10.4161/cc.5.18.3236. Epub 2006 Sep 15. Cell Cycle. 2006. PMID: 16969085 - Dbf4 is direct downstream target of ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related (ATR) protein to regulate intra-S-phase checkpoint.
Lee AY, Chiba T, Truong LN, Cheng AN, Do J, Cho MJ, Chen L, Wu X. Lee AY, et al. J Biol Chem. 2012 Jan 20;287(4):2531-43. doi: 10.1074/jbc.M111.291104. Epub 2011 Nov 28. J Biol Chem. 2012. PMID: 22123827 Free PMC article. - The Cdc7/Dbf4 protein kinase: target of the S phase checkpoint?
Jares P, Donaldson A, Blow JJ. Jares P, et al. EMBO Rep. 2000 Oct;1(4):319-22. doi: 10.1093/embo-reports/kvd076. EMBO Rep. 2000. PMID: 11269496 Free PMC article. Review. - The human intra-S checkpoint response to UVC-induced DNA damage.
Kaufmann WK. Kaufmann WK. Carcinogenesis. 2010 May;31(5):751-65. doi: 10.1093/carcin/bgp230. Epub 2009 Sep 30. Carcinogenesis. 2010. PMID: 19793801 Free PMC article. Review.
Cited by
- Clinical Candidates Targeting the ATR-CHK1-WEE1 Axis in Cancer.
Gorecki L, Andrs M, Korabecny J. Gorecki L, et al. Cancers (Basel). 2021 Feb 14;13(4):795. doi: 10.3390/cancers13040795. Cancers (Basel). 2021. PMID: 33672884 Free PMC article. Review. - c-Jun N-terminal kinase-mediated Rad18 phosphorylation facilitates Polη recruitment to stalled replication forks.
Barkley LR, Palle K, Durando M, Day TA, Gurkar A, Kakusho N, Li J, Masai H, Vaziri C. Barkley LR, et al. Mol Biol Cell. 2012 May;23(10):1943-54. doi: 10.1091/mbc.E11-10-0829. Epub 2012 Mar 28. Mol Biol Cell. 2012. PMID: 22456510 Free PMC article. - The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement.
Unsal-Kaçmaz K, Chastain PD, Qu PP, Minoo P, Cordeiro-Stone M, Sancar A, Kaufmann WK. Unsal-Kaçmaz K, et al. Mol Cell Biol. 2007 Apr;27(8):3131-42. doi: 10.1128/MCB.02190-06. Epub 2007 Feb 12. Mol Cell Biol. 2007. PMID: 17296725 Free PMC article. - Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation.
Zegerman P, Diffley JF. Zegerman P, et al. Nature. 2010 Sep 23;467(7314):474-8. doi: 10.1038/nature09373. Epub 2010 Sep 12. Nature. 2010. PMID: 20835227 Free PMC article. - Human papilloma virus type16 E6 deregulates CHK1 and sensitizes human fibroblasts to environmental carcinogens independently of its effect on p53.
Chen B, Simpson DA, Zhou Y, Mitra A, Mitchell DL, Cordeiro-Stone M, Kaufmann WK. Chen B, et al. Cell Cycle. 2009 Jun 1;8(11):1775-87. doi: 10.4161/cc.8.11.8724. Epub 2009 Jun 10. Cell Cycle. 2009. PMID: 19411857 Free PMC article.
References
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Annu Rev Biochem. 2004;73:39–85. - PubMed
- Dutta A, Bell SP. Annu Rev Cell Dev Biol. 1997;13:293–332. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- P30 ES010126/ES/NIEHS NIH HHS/United States
- CA055065/CA/NCI NIH HHS/United States
- R01 CA055065/CA/NCI NIH HHS/United States
- P30 CA016086/CA/NCI NIH HHS/United States
- T32 ES007017/ES/NIEHS NIH HHS/United States
- R01 ES011012/ES/NIEHS NIH HHS/United States
- ES07017/ES/NIEHS NIH HHS/United States
- ES11012/ES/NIEHS NIH HHS/United States
- P30-ES10126/ES/NIEHS NIH HHS/United States
- P30-CA 16086/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous