Cooperative roles of vertebrate Fbh1 and Blm DNA helicases in avoidance of crossovers during recombination initiated by replication fork collapse - PubMed (original) (raw)
Cooperative roles of vertebrate Fbh1 and Blm DNA helicases in avoidance of crossovers during recombination initiated by replication fork collapse
Masaoki Kohzaki et al. Mol Cell Biol. 2007 Apr.
Abstract
Fbh1 (F-box DNA helicase 1) orthologues are conserved from Schizosaccharomyces pombe to chickens and humans. Here, we report the disruption of the FBH1 gene in DT40 cells. Although the yeast fbh1 mutant shows an increase in sensitivity to DNA damaging agents, FBH1(-)(/)(-) DT40 clones show no prominent sensitivity, suggesting that the loss of FBH1 might be compensated by other genes. However, FBH1(-)(/)(-) cells exhibit increases in both sister chromatid exchange and the formation of radial structures between homologous chromosomes without showing a defect in homologous recombination. This phenotype is reminiscent of BLM(-)(/)(-) cells and suggests that Fbh1 may be involved in preventing extensive strand exchange during homologous recombination. In addition, disruption of RAD54, a major homologous recombination factor in FBH1(-)(/)(-) cells, results in a marked increase in chromosome-type breaks (breaks on both sister chromatids at the same place) following replication fork arrest. Further, FBH1BLM cells showed additive increases in both sister chromatid exchange and the formation of radial chromosomes. These data suggest that Fbh1 acts in parallel with Bloom helicase to control recombination-mediated double-strand-break repair at replication blocks and to reduce the frequency of crossover.
Figures
FIG. 1.
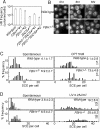
Gene targeting of the FBH1 locus. (A) Schematic representation of a part of the FBH1 locus and the targeting constructs. The FBH1 replacement constructs, which contain a bsr or hisD selection marker gene, were used to disrupt the FBH1 locus. The filled box represents the FBH1 exons. RI, EcoRI. (B) Southern blot analysis of EcoRI-digested DNA using the probe indicated in panel A. (C) RT-PCR analysis of total RNA using the primers shown in panel A. The coding region of the chicken β-actin gene was amplified as a control. (D) Normal growth kinetics of FBH1_−/_− cells. (E) Cell cycle distribution of the indicated cell lines. Cells were stained with FITC-anti-BrdU antibody (y axis, log scale) to detect BrdU uptake and with propidium iodide to measure the total DNA (x axis, linear scale). The upper gate identifies cells incorporating BrdU (∼S phase), the lower left gate identifies G1 cells, and the lower right gate displays G2/M cells. The sub-G1 fraction reflects dead cells. The numbers given in the boxes indicate the percentages of gated events.
FIG. 2.
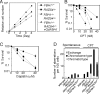
Sensitivities of the FBH1 mutant to genotoxic stresses. The indicated genotypes of cells were exposed to γ-rays (A) and UV (B), after a 1-h treatment with MMS (C) and cisplatin (D) and after continuous exposure to camptothecin (CPT) (E). The doses of genotoxic agents are displayed on the x axes on a linear scale, while the fractions of surviving colonies are displayed on the y axes on a logarithmic scale. Error bars show the standard error of the mean for at least three independent experiments.
FIG. 3.
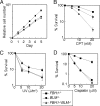
No significant defect in HR in FBH1_−/_− cells. (A) I-SceI-induced gene conversion assay with the indicated genotypes. The frequency of HR-dependent double-strand-break repair is shown as the number of G418-resistant colonies per 5 × 106 cells (white bars). The experiments were done at least three times. (B) Rad51 focus formation with the indicated genotypes before and at 3 and 6 h after 4 Gy γ-irradiation. (C) Spontaneous and 5 nM camptothecin (CPT)-induced SCE events in the macrochromosomes of 50 metaphase cells. Means ± standard errors are shown in the upper right corner. Underlined numbers indicate subtraction of spontaneous SCE from SCE following CPT treatment. The level of spontaneous SCE in FBH1_−/_− cells significantly differs from that in wild-type cells (P < 0.0001). The level of CPT-induced SCE was indistinguishable between wild-type and FBH1_−/_− cells (P < 0.0001). Statistical significance was calculated by the Mann-Whitney nonparametric U test (36). (D) UV-induced SCE (0.25 J/m2) is shown as in panel C. The level of UV-induced SCE was indistinguishable between wild-type and FBH1_−/_− cells (P < 0.0004).
FIG. 4.
FBH1_−/_− RAD54_−/_− cells display genome instability and hypersensitivity against camptothecin (CPT). (A) Growth curves of cells of the indicated genotypes. Results were obtained from at least three independent experiments. (B and C) Colony survival assay after treatment with CPT and cisplatin performed as described for Fig. 2. Error bars show the standard error of the mean for at least three independent experiments. (D) Chromosome aberrations before and following 10 nM CPT treatment for 9 h with the indicated genotypes.
FIG. 5.
Combined mutations of the FBH1 and BLM genes synergistically elevate sensitivity to camptothecin (CPT). (A) Growth curves of cells of the indicated genotypes. (B to D) Colony survival assay after treatment with CPT, UV, and cisplatin performed as described for Fig. 2. Error bars show the standard error of the mean for at least three independent experiments.
FIG. 6.
Increased genome instability of FBH1_−/_− BLM_−/_− cells observed in both UV-induced SCE and exchange between homologous chromosomes. Cells were labeled with BrdU during two cell cycle periods with (B) or without (A) prior UV treatment (0.25 J/m2). Spontaneous and UV-induced SCEs in the macrochromosomes of 50 metaphase cells were counted. Underlined numbers indicate subtraction of spontaneous SCE from SCE following UV. Histograms show the distribution of cells with the indicated numbers of SCEs per cell. The mean number of SCEs per cell ± standard error is shown in each histogram. The level of spontaneous SCE in BLM_−/_− cells significantly differs from that in FBH1_−/_− BLM_−/_− cells (P < 0.02). The level of UV-induced SCE was distinguishable between BLM_−/_− and FBH1_−/_− BLM_−/_− cells (P < 0.03), though it may have reached a plateau level. Statistical significance was calculated by the Mann-Whitney nonparametric U test. (C) Representative homologous chromosome exchange in FBH1_−/_− BLM_−/_− cells. Arrowheads indicate chromosome 1. (D) Example of nonhomologous exchange events between chromosomes 1 and 2 and between chromosomes 2 and 4 shown by arrowheads. (E) Frequency of chromosomal aberrations in mitotic cells harvested at indicated times after exposure of asynchronous cell populations to 1 J/m2 UV. Chromosome aberrations were scored for 100 metaphase cells. All samples were treated with Colcemid for the last 3 h prior to harvest of cells.
FIG. 7.
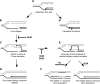
Model for the function of Fbh1 and Blm at stalled replication forks. A replication fork encounters damages on the template (oval) and is blocked. This block can be bypassed by translesion synthesis (transition from A to G), or the fork can collapse (transition from A to B). HR to restart the collapsed fork is usually carried out by synthesis-dependent strand annealing (SDSA), which is not associated with crossover (transition from C to D). More extensive strand exchange between sister chromatids (E) leads to the formation of a double Holliday junction, which may be resolved with crossover (F). Blm may play a number of roles that reduce crossovers. It may suppress D-loop formation (transition from B to C) (40), reverse Holliday junction formation (transition from C to E) (26) and, with topoisomerase IIIα, promote the dissolution of double Holliday junctions, thereby avoiding resolution by incision (42). The present data support a role for Fbh1 acting alongside Blm in the second of these possibilities, the avoidance of extensive strand exchange and Holliday junction formation (transition from C to E).
Similar articles
- Possible association of BLM in decreasing DNA double strand breaks during DNA replication.
Wang W, Seki M, Narita Y, Sonoda E, Takeda S, Yamada K, Masuko T, Katada T, Enomoto T. Wang W, et al. EMBO J. 2000 Jul 3;19(13):3428-35. doi: 10.1093/emboj/19.13.3428. EMBO J. 2000. PMID: 10880455 Free PMC article. - Role of the Schizosaccharomyces pombe F-Box DNA helicase in processing recombination intermediates.
Morishita T, Furukawa F, Sakaguchi C, Toda T, Carr AM, Iwasaki H, Shinagawa H. Morishita T, et al. Mol Cell Biol. 2005 Sep;25(18):8074-83. doi: 10.1128/MCB.25.18.8074-8083.2005. Mol Cell Biol. 2005. PMID: 16135799 Free PMC article. - The absence of a functional relationship between ATM and BLM, the components of BASC, in DT40 cells.
Wang W, Seki M, Otsuki M, Tada S, Takao N, Yamamoto K, Hayashi M, Honma M, Enomoto T. Wang W, et al. Biochim Biophys Acta. 2004 Mar 2;1688(2):137-44. doi: 10.1016/j.bbadis.2003.11.008. Biochim Biophys Acta. 2004. PMID: 14990344 - Role of the BLM helicase in replication fork management.
Wu L. Wu L. DNA Repair (Amst). 2007 Jul 1;6(7):936-44. doi: 10.1016/j.dnarep.2007.02.007. Epub 2007 Mar 23. DNA Repair (Amst). 2007. PMID: 17363339 Review.
Cited by
- FBH1 helicase disrupts RAD51 filaments in vitro and modulates homologous recombination in mammalian cells.
Simandlova J, Zagelbaum J, Payne MJ, Chu WK, Shevelev I, Hanada K, Chatterjee S, Reid DA, Liu Y, Janscak P, Rothenberg E, Hickson ID. Simandlova J, et al. J Biol Chem. 2013 Nov 22;288(47):34168-34180. doi: 10.1074/jbc.M113.484493. Epub 2013 Oct 9. J Biol Chem. 2013. PMID: 24108124 Free PMC article. - Repair Pathway Choices and Consequences at the Double-Strand Break.
Ceccaldi R, Rondinelli B, D'Andrea AD. Ceccaldi R, et al. Trends Cell Biol. 2016 Jan;26(1):52-64. doi: 10.1016/j.tcb.2015.07.009. Epub 2015 Oct 1. Trends Cell Biol. 2016. PMID: 26437586 Free PMC article. Review. - Hitting the bull's eye: novel directed cancer therapy through helicase-targeted synthetic lethality.
Aggarwal M, Brosh RM Jr. Aggarwal M, et al. J Cell Biochem. 2009 Apr 1;106(5):758-63. doi: 10.1002/jcb.22048. J Cell Biochem. 2009. PMID: 19173305 Free PMC article. Review. - RECQL5/Recql5 helicase regulates homologous recombination and suppresses tumor formation via disruption of Rad51 presynaptic filaments.
Hu Y, Raynard S, Sehorn MG, Lu X, Bussen W, Zheng L, Stark JM, Barnes EL, Chi P, Janscak P, Jasin M, Vogel H, Sung P, Luo G. Hu Y, et al. Genes Dev. 2007 Dec 1;21(23):3073-84. doi: 10.1101/gad.1609107. Epub 2007 Nov 14. Genes Dev. 2007. PMID: 18003859 Free PMC article. - Srs2: the "Odd-Job Man" in DNA repair.
Marini V, Krejci L. Marini V, et al. DNA Repair (Amst). 2010 Mar 2;9(3):268-75. doi: 10.1016/j.dnarep.2010.01.007. Epub 2010 Jan 21. DNA Repair (Amst). 2010. PMID: 20096651 Free PMC article. Review.
References
- Adachi, N., S. So, and H. Koyama. 2004. Loss of nonhomologous end joining confers camptothecin resistance in DT40 cells. Implications for the repair of topoisomerase I-mediated DNA damage. J. Biol. Chem. 279:37343-37348. - PubMed
- Alexeev, A., A. Mazin, and S. C. Kowalczykowski. 2003. Rad54 protein possesses chromatin-remodeling activity stimulated by the Rad51-ssDNA nucleoprotein filament. Nat. Struct. Biol. 10:182-186. - PubMed
- Arnaudeau, C., C. Lundin, and T. Helleday. 2001. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 307:1235-1245. - PubMed
- Bezzubova, O., A. Silbergleit, Y. Yamaguchi-Iwai, S. Takeda, and J. M. Buerstedde. 1997. Reduced X-ray resistance and homologous recombination frequencies in a RAD54−/− mutant of the chicken DT40 cell line. Cell 89:185-193. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases