Preclinical validation of a multiplex real-time assay to quantify SMN mRNA in patients with SMA - PubMed (original) (raw)
Preclinical validation of a multiplex real-time assay to quantify SMN mRNA in patients with SMA
L R Simard et al. Neurology. 2007.
Abstract
Objective: To determine whether survival motor neuron (SMN) expression was stable over time.
Methods: We developed a multiplex real-time reverse transcriptase (RT)-PCR assay to quantify SMN transcripts in preclinical blood samples from 42 patients with spinal muscular atrophy (SMA) drawn for three time points per patient; most blood samples were shipped to a centralized laboratory.
Results: We obtained a sufficient amount (9.7 +/- 5.6 microg) of good-quality total RNA, and RNAs were stable for up to a 3-year interval. This allowed RNA samples collected during a 9- to 12-month period to be analyzed in a single run, thus minimizing interexperimental variability. SMN expression was stable over time; intersample variability for baseline measures, collected during a 17-month interval, was less than 15% for 38 of 42 SMA patients analyzed. This variability was well below the 1.95-fold increase in full-length SMN (flSMN) transcripts detected in SMA fibroblasts treated with 10 mM valproic acid.
Conclusion: Real-time quantification of SMN messenger RNA expression may be a biomarker that is amenable to multicenter SMA clinical trials.
Figures
Figure 1
Relationship between flSMN transcript quantity and SMN2 copy number. Quantification of flSMN was carried out by multiplex RT-PCR using PGK1 as an endogenous control. SMN2 copy number was evaluated as previously described.
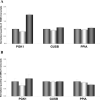
Figure 2
Effect of VPA treatment of type I SMA fibroblast cells. SMA type I fibroblast cell line 3831 was treated with 0, 1, or 10 mM VPA (medium, light, and dark gray cylinders, respectively), and the amount of flSMN (A) or Δ7SMN (B) transcripts was determined using either PGK1, GUSB, or PPIA as endogenous controls. Data are reported as means ± SEM of two experiments.
Similar articles
- A sensitive assay for measuring SMN mRNA levels in peripheral blood and in muscle samples of patients affected with spinal muscular atrophy.
Vezain M, Saugier-Veber P, Melki J, Toutain A, Bieth E, Husson M, Pedespan JM, Viollet L, Pénisson-Besnier I, Fehrenbach S, Bou J, Frébourg T, Tosi M. Vezain M, et al. Eur J Hum Genet. 2007 Oct;15(10):1054-62. doi: 10.1038/sj.ejhg.5201885. Epub 2007 Jul 4. Eur J Hum Genet. 2007. PMID: 17609673 - Hydroxyurea enhances SMN2 gene expression in spinal muscular atrophy cells.
Grzeschik SM, Ganta M, Prior TW, Heavlin WD, Wang CH. Grzeschik SM, et al. Ann Neurol. 2005 Aug;58(2):194-202. doi: 10.1002/ana.20548. Ann Neurol. 2005. PMID: 16049920 - [The expression of SMN2 gene mRNA in neuron-like cells derived from patients with spinal muscular atrophy].
Luo XM, Yang XS, Xiao B. Luo XM, et al. Zhonghua Nei Ke Za Zhi. 2006 Oct;45(10):831-4. Zhonghua Nei Ke Za Zhi. 2006. PMID: 17217749 Chinese. - Therapeutics development for spinal muscular atrophy.
Sumner CJ. Sumner CJ. NeuroRx. 2006 Apr;3(2):235-45. doi: 10.1016/j.nurx.2006.01.010. NeuroRx. 2006. PMID: 16554261 Free PMC article. Review. - Fishing for a mechanism: using zebrafish to understand spinal muscular atrophy.
Beattie CE, Carrel TL, McWhorter ML. Beattie CE, et al. J Child Neurol. 2007 Aug;22(8):995-1003. doi: 10.1177/0883073807305671. J Child Neurol. 2007. PMID: 17761655 Review.
Cited by
- Treatment of spinal muscular atrophy cells with drugs that upregulate SMN expression reveals inter- and intra-patient variability.
Also-Rallo E, Alías L, Martínez-Hernández R, Caselles L, Barceló MJ, Baiget M, Bernal S, Tizzano EF. Also-Rallo E, et al. Eur J Hum Genet. 2011 Oct;19(10):1059-65. doi: 10.1038/ejhg.2011.89. Epub 2011 May 25. Eur J Hum Genet. 2011. PMID: 21610752 Free PMC article. - Evaluation of SMN protein, transcript, and copy number in the biomarkers for spinal muscular atrophy (BforSMA) clinical study.
Crawford TO, Paushkin SV, Kobayashi DT, Forrest SJ, Joyce CL, Finkel RS, Kaufmann P, Swoboda KJ, Tiziano D, Lomastro R, Li RH, Trachtenberg FL, Plasterer T, Chen KS; Pilot Study of Biomarkers for Spinal Muscular Atrophy Trial Group. Crawford TO, et al. PLoS One. 2012;7(4):e33572. doi: 10.1371/journal.pone.0033572. Epub 2012 Apr 27. PLoS One. 2012. PMID: 22558076 Free PMC article. Clinical Trial. - SMA CARNI-VAL trial part I: double-blind, randomized, placebo-controlled trial of L-carnitine and valproic acid in spinal muscular atrophy.
Swoboda KJ, Scott CB, Crawford TO, Simard LR, Reyna SP, Krosschell KJ, Acsadi G, Elsheik B, Schroth MK, D'Anjou G, LaSalle B, Prior TW, Sorenson SL, Maczulski JA, Bromberg MB, Chan GM, Kissel JT; Project Cure Spinal Muscular Atrophy Investigators Network. Swoboda KJ, et al. PLoS One. 2010 Aug 19;5(8):e12140. doi: 10.1371/journal.pone.0012140. PLoS One. 2010. PMID: 20808854 Free PMC article. Clinical Trial. - Novel challenges in spinal muscular atrophy - How to screen and whom to treat?
Saffari A, Kölker S, Hoffmann GF, Weiler M, Ziegler A. Saffari A, et al. Ann Clin Transl Neurol. 2018 Nov 13;6(1):197-205. doi: 10.1002/acn3.689. eCollection 2019 Jan. Ann Clin Transl Neurol. 2018. PMID: 30656198 Free PMC article. Review. - Spinal muscular atrophy: advances in research and consensus on care of patients.
Wang CH, Lunn MR. Wang CH, et al. Curr Treat Options Neurol. 2008 Nov;10(6):420-8. doi: 10.1007/s11940-008-0044-7. Curr Treat Options Neurol. 2008. PMID: 18990310
References
- Crawford TO, Pardo CA. The neurobiology of childhood spinal muscular atrophy. Neurobiol Dis. 1996;3:97–110. - PubMed
- Lefebvre S, Burglen L, Reboullet S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. - PubMed
- Wirth B. An update of the mutation spectrum of the survival motor neuron gene (SMN1) in autosomal recessive spinal muscular atrophy (SMA). Hum Mutat. 2000;15:228–237. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials