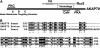Ssh4, Rcr2 and Rcr1 affect plasma membrane transporter activity in Saccharomyces cerevisiae - PubMed (original) (raw)
Ssh4, Rcr2 and Rcr1 affect plasma membrane transporter activity in Saccharomyces cerevisiae
Jhansi Kota et al. Genetics. 2007 Apr.
Abstract
Nutrient uptake in the yeast Saccharomyces cerevisiae is a highly regulated process. Cells adjust levels of nutrient transporters within the plasma membrane at multiple stages of the secretory and endosomal pathways. In the absence of the ER-membrane-localized chaperone Shr3, amino acid permeases (AAP) inefficiently fold and are largely retained in the ER. Consequently, shr3 null mutants exhibit greatly reduced rates of amino acid uptake due to lower levels of AAPs in their plasma membranes. To further our understanding of mechanisms affecting AAP localization, we identified SSH4 and RCR2 as high-copy suppressors of shr3 null mutations. The overexpression of SSH4, RCR2, or the RCR2 homolog RCR1 increases steady-state AAP levels, whereas the genetic inactivation of these genes reduces steady-state AAP levels. Additionally, the overexpression of any of these suppressor genes exerts a positive effect on phosphate and uracil uptake systems. Ssh4 and Rcr2 primarily localize to structures associated with the vacuole; however, Rcr2 also localizes to endosome-like vesicles. Our findings are consistent with a model in which Ssh4, Rcr2, and presumably Rcr1, function within the endosome-vacuole trafficking pathway, where they affect events that determine whether plasma membrane proteins are degraded or routed to the plasma membrane.
Figures
Figure 1.—
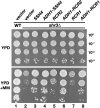
SSH4, RCR2, and RCR1 are high-copy suppressors of shr3 deletion. Tenfold dilution series of cell suspensions of CAY28 (wild type) transformed with pRS202 (vector) and JKY1 (_shr3_Δ) transformed with 2μ plasmids pRS202 (vector), pJK120 (SSH4), pJK123 (_ADH1_-SSH4), pJK121 (RCR2), pJK124 (P_ADH1_-RCR2), pJK122 (RCR1), or pJK125 (_ADH1_-RCR1) were spotted onto solid YPD medium and YPD supplemented with MM. Plates were incubated at 30° for 2 days (YPD) or for 3 days (YPD+MM) and photographed.
Figure 2.—
Suppression of _shr3_Δ by SSH4, RCR2, and RCR1 is not due to chaperone-like activity in the ER. (A) Suppression of _shr3_Δ on
d
-histidine-containing medium. Serial dilutions of cell suspensions of wild-type strain (CAY28) carrying vector control (pRS202) and _shr3_Δ strain (JKY1) carrying vector control (pRS202) or plasmid-expressing SSH4 (pJK123), RCR2 (pJK124), or RCR1 (pJK125) from the ADH1 promoter were prepared. Equal aliquots of each dilution were applied to SAD or SAD supplemented with
d
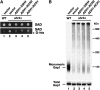
-histidine (0.15%). Plates were incubated at 30° for 2 days and photographed. (B) Analysis of Gap1 aggregation. Protein extracts from strains as in A grown in SAD were prepared and solubilized with DM (1.5 μg DM μg−1 protein) at 4°. Solubilized proteins were separated by BN–PAGE, immunoblotted, and analyzed using antibodies raised against Gap1 (top). Total Gap1 levels were analyzed in protein extracts (20 μg) separated by SDS–PAGE (bottom).
Figure 3.—
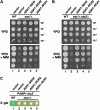
SSH4, RCR2, and RCR1 suppress loss of SPS sensor signaling. (A) SSH4, RCR2, and RCR1 are high-copy suppressors of _ssy1_Δ. Phenotypic analysis of wild-type (CAY28) and _ssy1_Δ strains (CAY91) carrying a vector control (pRS202) or expressing SSH4 (pJK123), RCR2 (pJK124), or RCR1 (pJK125) as indicated. Tenfold dilution series of cell suspensions were spotted onto plates containing YPD and YPD supplemented with MM. Plates were incubated at 30° for 2 days (YPD) or for 4 days (YPD+MM) and photographed. (B) SSH4, RCR2, and RCR1 are high-copy suppressors of _stp1_Δ _stp2_Δ. Serial dilutions of wild-type (CAY28) and _stp1_Δ stp2_Δ strains (CAY91) carrying a vector control (pRS202) or plasmids expressing SSH4 (pJK123), RCR2 (pJK124), or RCR1 (pJK125) are as indicated. Dilutions of cell suspensions were applied to YPD or YPD with MM as indicated. Plates were incubated as in A and photographed. (C) Expression from the AGP1 promoter was monitored by assessing β-galactosidase activity in cells transformed with a PAGP1-lacZ reporter construct. Wild-type strain PLY126 cotransformed with pCA030 (YCp_AGP1-lacZ) and pRS202 (vector), as well as _ssy1_Δ strain HKY20 carrying pCA030 together with 2μ plasmids expressing SSH4 (pJK123), RCR2 (pJK124), or RCR1 (pJK125) from the ADH1 promoter were applied to solid SD medium supplemented with 1.3 m
m
leucine. Plates were grown at 30° for 2 days and overlaid with X-gal substrate.
Figure 4.—
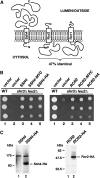
SSH4, RCR2, and RCR1 encode membrane proteins. (A) Schematic of secondary structure predictions of Ssh4, Rcr2, and Rcr1. Hydropathy analysis using a window size of 11 amino acid residues indicates that all three proteins harbor a single membrane-spanning segment in close proximity to their N termini (K
yte
and D
oolittle
1982). The orientation in the membrane is based on the N-best algorithm (TMHMM 1.0 prediction). (B) Functional analysis of epitope-tagged SSH4 and RCR2. Serial dilutions of strain HFY501 (WT: leu2 SHR3) transformed with pRS202 (vector) and strain HFY500 (_leu2 shr3_Δ) transformed with 2μ plasmids pRS202 (vector), pJK120 (SSH4), pJK131 (SSH4-3xMYC), pJK129 (SSH4-6xHA), pJK121 (RCR2), pJK135 (RCR2-3xMYC), or pJK133 (RCR2-6xHA) were applied to SC (−ura) plates. Plates were incubated at 30° for 3 days and photographed. (C) Immunoblot analysis of whole-cell extracts from strain JKY40 (_ssh4_Δ) transformed with either pJK120 (SSH4) or pJK130 (SSH4-6xHA) and strain JKY41 (_rcr2_Δ) transformed with either pJK121 (RCR2) or pJK134 (RCR2-6xHA), grown to log phase in SC (−ura) medium. Antibodies used to visualize Ssh4-HA and Rcr2-HA were α-HA (12CA5, mouse) and α-HA (3E10, rat), respectively.
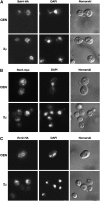
Figure 5.—
Localization of Ssh4 and Rcr2. (A) Indirect immunolocalization of Ssh4-HA in strain JKY40 (_ssh4_Δ) transformed with SSH4-6xHA expressed from either a CEN plasmid (pJK130, top) or a 2μ plasmid (pJK129, bottom). (B) Rcr2-myc was localized in JKY41 (_rcr2_Δ) transformed with either RCR2-3xMYC expressed from a CEN plasmid (pJK136, top) or from a 2μ plasmid (pJK135, bottom). (C) Indirect immunolocalization of Rcr2-HA was performed with RCR2-6xHA expressed from either a CEN plasmid (pJK134, top) or a 2μ plasmid (pJK133, bottom) in JKY41 (_rcr2_Δ). Cells expressing Ssh4-HA, Rcr2-myc, or Rcr2-HA were processed for indirect immunofluorescence and viewed by Alexa Fluor 488-dependent fluorescence, DAPI staining, and Nomarski optics as indicated.
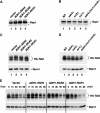
Figure 6.—
Analysis of Gap1 and Tat2 protein levels. (A) Gap1 levels in whole-cell extracts from a wild-type strain (CAY28) transformed with pRS202 (vector), pJK123 (P_ADH1-SSH4_), pJK124 (P_ADH1-RCR2_), or pJK125 (P_ADH1-RCR1_). Cells were grown to an OD600 of 1.5 in SAD, and proteins in cell lysates were resolved by SDS–PAGE and immunoblotted with α-Gap1 antibodies. The asterisk indicates a nonspecific immunoreactive band unrelated to Gap1 that serves as a loading control. (B) Gap1 levels in whole-cell extracts from wild-type (CAY28), _ssh4_Δ (JKY40), _rcr2_Δ (JKY41), _rcr1_Δ (JKY42), and ssh4_Δ rcr2_Δ rcr1_Δ (HFY538) strains. Cells were grown as in SAD supplemented with uracil, and extracts were prepared and analyzed as in A. (C) Steady-state levels of HA-TAT2 were analyzed in strain PLY860 cotransformed with pJK139 (HA-TAT2) and 2μ plasmid pRS202 (vector), pJK123 (P_ADH1-SSH4), pJK124 (P_ADH1-RCR2), or pJK125 (P_ADH1-RCR1) grown in SC (−ura,−trp) to an OD600 of 2. Cell extracts were resolved by SDS–PAGE and immunoblotted with α-HA (3F10, rat) and α-Dpm1 antibodies. (D) Immunoblotting of extracts from wild-type strain (CAY28), _ssh4_Δ (JKY40), _rcr2_Δ (JKY41), _rcr1_Δ (JKY42), and _ssh4_Δ _rcr2_Δ _rcr1_Δ (HFY538) transformed with plasmid pAS55 (HA-TAT2). Cells were grown SC (−ura) to an OD600 of 2 and proteins were resolved and analyzed as in C. (E) Strain PLY860 cotransformed with plasmids as in C was grown in SC (−ura −trp) to an OD600 of 2. Rapamycin was added and whole-cell extracts were prepared at the time points indicated. Proteins were resolved and analyzed as in C.
Figure 7.—
Overexpression of SSH4, RCR2, and RCR1 affects multiple plasma membrane transport systems. (A) Suppression of _pho86_Δ. Serial dilutions of PHO86 (CEN.PK113-5D/PHO84myc) and pho86_Δ strains (JKY6) carrying pRS202 (vector) or plasmids expressing SSH4 (pJK123), RCR2 (pJK124), or RCR1 (pJK125) from the P_ADH1 promoter, as indicated, were spotted onto high-phosphate (YPD) and low-phosphate (SD 0.2 m
m
phosphate) media. Plates were incubated at 30° for 3 days and photographed. (B) Growth characteristics on FOA-containing media. Wild-type strain CAY28 was transformed with 2μ plasmids pRS202 (vector), pJK120 (SSH4), pJK123 (_ADH1_-SSH4), pJK121 (RCR2), pJK124 (_ADH1_-RCR2), pJK122 (RCR1), or pJK125 (_ADH1_-RCR1) and 10-fold dilution series were spotted onto SD and SD+FOA (0.1 mg/ml). Plates were incubated at 30° for 3 days and photographed.
Figure 8.—
The C-terminal region of Rcr2 shows homology to the human scaffold AKAP79. (A) Schematic of the structural features of yeast Rcr2 and human AKAP79. The predicted transmembrane spanning domain (TM) of Rcr2: basic membrane-targeting domains (A–C), PKC, CaN, and cAMP-dependent kinase (PKA) binding regions of AKAP79 are indicated. Sequence homology of Rcr2 and AKAP79 was found in the region indicated between the dashed lines. (B) Multiple alignment of human AKAP79 (aa 292–381), bovine AKAP75 (aa 293–382), and yeast Rcr2 (aa 89–198) sequences (CLUSTAL W, Biology WorkBench 3.2, San Diego Supercomputer Center). Amino acids that are identical in all three proteins (white lettering, solid boxes), as well as amino acids with high similarity (white lettering, darkly shaded boxes) and low similarity (black lettering, lightly shaded boxes) are indicated.
Similar articles
- Membrane chaperone Shr3 assists in folding amino acid permeases preventing precocious ERAD.
Kota J, Gilstring CF, Ljungdahl PO. Kota J, et al. J Cell Biol. 2007 Feb 26;176(5):617-28. doi: 10.1083/jcb.200612100. J Cell Biol. 2007. PMID: 17325204 Free PMC article. - SHR3: a novel component of the secretory pathway specifically required for localization of amino acid permeases in yeast.
Ljungdahl PO, Gimeno CJ, Styles CA, Fink GR. Ljungdahl PO, et al. Cell. 1992 Oct 30;71(3):463-78. doi: 10.1016/0092-8674(92)90515-e. Cell. 1992. PMID: 1423607 - Shr3p mediates specific COPII coatomer-cargo interactions required for the packaging of amino acid permeases into ER-derived transport vesicles.
Gilstring CF, Melin-Larsson M, Ljungdahl PO. Gilstring CF, et al. Mol Biol Cell. 1999 Nov;10(11):3549-65. doi: 10.1091/mbc.10.11.3549. Mol Biol Cell. 1999. PMID: 10564255 Free PMC article. - Protein transport from the late Golgi to the vacuole in the yeast Saccharomyces cerevisiae.
Bowers K, Stevens TH. Bowers K, et al. Biochim Biophys Acta. 2005 Jul 10;1744(3):438-54. doi: 10.1016/j.bbamcr.2005.04.004. Biochim Biophys Acta. 2005. PMID: 15913810 Review. - The ubiquitin code of yeast permease trafficking.
Lauwers E, Erpapazoglou Z, Haguenauer-Tsapis R, André B. Lauwers E, et al. Trends Cell Biol. 2010 Apr;20(4):196-204. doi: 10.1016/j.tcb.2010.01.004. Trends Cell Biol. 2010. PMID: 20138522 Review.
Cited by
- Ear1p and Ssh4p are new adaptors of the ubiquitin ligase Rsp5p for cargo ubiquitylation and sorting at multivesicular bodies.
Léon S, Erpapazoglou Z, Haguenauer-Tsapis R. Léon S, et al. Mol Biol Cell. 2008 Jun;19(6):2379-88. doi: 10.1091/mbc.e08-01-0068. Epub 2008 Mar 26. Mol Biol Cell. 2008. PMID: 18367543 Free PMC article. - A Cycle of Ubiquitination Regulates Adaptor Function of the Nedd4-Family Ubiquitin Ligase Rsp5.
MacDonald C, Shields SB, Williams CA, Winistorfer S, Piper RC. MacDonald C, et al. Curr Biol. 2020 Feb 3;30(3):465-479.e5. doi: 10.1016/j.cub.2019.11.086. Epub 2020 Jan 16. Curr Biol. 2020. PMID: 31956026 Free PMC article. - Arrestin-mediated endocytosis of yeast plasma membrane transporters.
Nikko E, Pelham HR. Nikko E, et al. Traffic. 2009 Dec;10(12):1856-67. doi: 10.1111/j.1600-0854.2009.00990.x. Traffic. 2009. PMID: 19912579 Free PMC article. - Out-of-the-groove transport of lipids by TMEM16 and GPCR scramblases.
Malvezzi M, Andra KK, Pandey K, Lee BC, Falzone ME, Brown A, Iqbal R, Menon AK, Accardi A. Malvezzi M, et al. Proc Natl Acad Sci U S A. 2018 Jul 24;115(30):E7033-E7042. doi: 10.1073/pnas.1806721115. Epub 2018 Jun 20. Proc Natl Acad Sci U S A. 2018. PMID: 29925604 Free PMC article. - The WAGR syndrome gene PRRG4 is a functional homologue of the commissureless axon guidance gene.
Justice ED, Barnum SJ, Kidd T. Justice ED, et al. PLoS Genet. 2017 Aug 31;13(8):e1006865. doi: 10.1371/journal.pgen.1006865. eCollection 2017 Aug. PLoS Genet. 2017. PMID: 28859078 Free PMC article.
References
- Bartnicki-Garcia, S., 2006. Chitosomes: past, present and future. FEMS Yeast Res. 6: 957–965. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous