An African origin for the intimate association between humans and Helicobacter pylori - PubMed (original) (raw)
. 2007 Feb 22;445(7130):915-918.
doi: 10.1038/nature05562. Epub 2007 Feb 7.
François Balloux # 2, Yoshan Moodley 1, Andrea Manica 3, Hua Liu 2, Philippe Roumagnac 1, Daniel Falush 4, Christiana Stamer 1, Franck Prugnolle 5, Schalk W van der Merwe 6, Yoshio Yamaoka 7, David Y Graham 7, Emilio Perez-Trallero 8, Torkel Wadstrom 9, Sebastian Suerbaum 10, Mark Achtman 1
Affiliations
- PMID: 17287725
- PMCID: PMC1847463
- DOI: 10.1038/nature05562
An African origin for the intimate association between humans and Helicobacter pylori
Bodo Linz et al. Nature. 2007.
Abstract
Infection of the stomach by Helicobacter pylori is ubiquitous among humans. However, although H. pylori strains from different geographic areas are associated with clear phylogeographic differentiation, the age of an association between these bacteria with humans remains highly controversial. Here we show, using sequences from a large data set of bacterial strains that, as in humans, genetic diversity in H. pylori decreases with geographic distance from east Africa, the cradle of modern humans. We also observe similar clines of genetic isolation by distance (IBD) for both H. pylori and its human host at a worldwide scale. Like humans, simulations indicate that H. pylori seems to have spread from east Africa around 58,000 yr ago. Even at more restricted geographic scales, where IBD tends to become blurred, principal component clines in H. pylori from Europe strongly resemble the classical clines for Europeans described by Cavalli-Sforza and colleagues. Taken together, our results establish that anatomically modern humans were already infected by H. pylori before their migrations from Africa and demonstrate that H. pylori has remained intimately associated with their human host populations ever since.
Figures
FIGURE 1
Five ancestral populations in H. pylori. a) DISTRUCT plot of the proportions of ancestral nucleotides in 769 H. pylori isolates as determined by Structure V2.0 (linkage model). A thin line for each isolate indicates the estimated amount of ancestry from each ancestral population as five colored segments. The lines are grouped by (sub)population and ordered by ancestry. b) Neighbor joining tree (black lines) of the relationships between and diversity within five ancestral populations calculated as described. Circle diameters are proportional to within-population genetic diversity and angles of filled arcs are proportional to the amount of ancestry attributable to each population among modern strains. scale bar: 0.01. c-g) Spatial distribution of five geographic sources of ancestral nucleotides. Dark-light gradients show clinal declines in proportions of ancestral nucleotides by distance from a geographic centre. Color-coding as in part b. AE1, AE2: ancestral Europe 1 and 2.
FIGURE 2
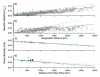
Parallel geographic patterns of genetic diversity in humans (a,c) and H. pylori (b, d). a, b) Genetic distance between pairs of geographic populations (_F_ST) versus geographic distance between the two populations. c, d) Average gene diversity within geographic populations (HS) versus geographic distance from East Africa. R2: a,0.77; b,0.47; c,0.85; d,0.59. In part d, samples that are predominantly composed of hpEurope isolates are indicated by filled circles while the red circle identifies the sample from South Africa. Confidence intervals are indicated by dotted lines.
FIGURE 3
Similar clinal gradients between principal components 1-3 from European H. pylori (a-c, concatenated sequences) and humans (d-f, allozymes). Figs d-f, reprinted with permission from citation 7(copyright 1995 National Academy of Sciences, U.S.A.).
References
- Achtman M, et al. Recombination and clonal groupings within Helicobacter pylori from different geographical regions. Mol. Microbiol. 1999;32:459–470. - PubMed
- Falush D, et al. Traces of human migrations in Helicobacter pylori populations. Science. 2003;299:1582–1585. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical