Structural basis for nicotinamide inhibition and base exchange in Sir2 enzymes - PubMed (original) (raw)
Structural basis for nicotinamide inhibition and base exchange in Sir2 enzymes
Brandi D Sanders et al. Mol Cell. 2007.
Abstract
The Sir2 family of proteins consists of broadly conserved NAD(+)-dependent deacetylases that are implicated in diverse biological processes, including DNA regulation, metabolism, and longevity. Sir2 proteins are regulated in part by the cellular concentrations of a noncompetitive inhibitor, nicotinamide, that reacts with a Sir2 reaction intermediate via a base-exchange reaction to reform NAD(+) at the expense of deacetylation. To gain a mechanistic understanding of nicotinamide inhibition in Sir2 enzymes, we captured the structure of nicotinamide bound to a Sir2 homolog, yeast Hst2, in complex with its acetyl-lysine 16 histone H4 substrate and a reaction intermediate analog, ADP-HPD. Together with related biochemical studies and structures, we identify a nicotinamide inhibition and base-exchange site that is distinct from the so-called "C pocket" binding site for the nicotinamide group of NAD(+). These results provide insights into the Sir2 mechanism of nicotinamide inhibition and have important implications for the development of Sir2-specific effectors.
Figures
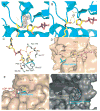
Figure 1. Structures of the free yHst2/ADP-HPD/histone H4 complex
(A) Ternary yHst2 (gray) complex, highlighting strictly conserved (pink) and conserved (red) residues, the binding sites of acetyl-lysine (green), carba-NAD+ (cyan) and ADP-ribose (yellow) and the conserved C and D pockets. Hydrogen bonds between the acetyl-lysine and carba-NAD+ are shown as yellow dotted lines. Residues 43–48 of the flexible loop and residue 64 were omitted for clarity. (B) Superimposition of the yHst2/ADP-ribose/H4 complex (magenta) with the yHst2/ADP-HPD/H4 complex (cyan) and the yHst2/ADP-HPD/H4 complex bound to nicotinamide (blue). The intermediate analogue, acetylated histone H4 ligands, and nicotinamide are shown in green for the ADP-ribose complex, yellow for the free ADP-HPD complex, and orange for the nicotinamide bound ADP-HDP complex. (C) Simulated annealing omit density contoured at 1.0 sigma showing density for the protein (blue) and ADP-HPD (atoms individually colored). Water molecules are shown as blue spheres. (D) yHst2 bound to ADP-HPD (atoms individually colored) and highlighting residues that make hydrogen bonds (red dashed lines) or van der Waals contacts with ADP-HPD. Hydrogen bonding residues are colored pink, residues that make van der Waals interactions are colored cyan, and residues that make both interactions are colored purple.
Figure 2. The nicotinamide binding pocket
(A) Fo-Fc map contoured at 2.0 sigma (blue) prior to the addition of nicotinamide for protein refinement, and simulated annealing omit density contoured at 1.0 sigma (pink) showing nicotinamide density for the ternary yHst2/ADP-HPD/H4 complex soaked with nicotinamide. The protein is shown in blue and the acetyl-lysine, ADP-HPD and nicotinamide atoms are shown individually colored. (B) Corresponding 2 sigma Fo-Fc map for the unsoaked crystals. (C) Interactions between nicotinamide (atoms individually colored), yHst2 (white backbone) and acetyl-lysine 16 of histone H4 (green backbone). Residues that make van der Waals interactions and hydrogen bonds (red) to nicotinamide are shown. (D) yHst2 bound to nicotinamide, acetyllysine, and ADP-HPD highlighting residues that make van der Waals or hydrogen bonding interactions with nicotinamide. (E) yHst2 in a view roughly orthogonal to and with the same color coding as (D), but highlighting residues that form the tunnel (red) through which nicotinamide is proposed to diffuse. (F) yHst2 (gray) shown in surface representation with tunnel residues from the yHst2/carba-NAD+ (pink), yHst2/2′-O-acetyl ADP-ribose/Acetyl-lysine (cyan), yHst2/carba-NAD+/Acetyl-lysine (yellow) and yHst2/ADP-HPD/Acetyl-lysine (purple) structures highlighted.
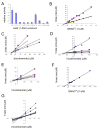
Figure 3. Activity, kinetic, and inhibition data for wild-type and mutant yHst2
(A) Relative activity of wild-type and mutant yHst2 enzymes based on their ability to deacetylate a fluorescently-labeled acetylated peptide in duplicate. Error bars represent one SD of experiments done at least in triplicate. (B) Lineweaver-Burk plot describing the NAD+ binding kinetics of wild-type yHst2 (blue diamonds), the yHst2 I117F and I117V mutants (pink squares, yellow triangles, respectively) done in triplicate. (C) Dixon plot describing the nicotinamide inhibition of wild-type yHst2 at varying concentrations of NAD+, 15 μM (pink squares), 25 μM (yellow triangles), 50 μM (cyan diamonds), and 80 μM (purple circles). Each line is fit to an equation for a noncompetitive inhibitor with obtained in triplicate. (D) Dixon plot describing the nicotinamide inhibition of yHst2 I117F with conditions as described in (C), but also including 5 μM NAD+ (blue diamonds). (E) Dixon plot describing the nicotinamide inhibition of yHst2 I117V with conditions as described in (D). (F) Lineweaver-Burk plot describing the NAD+ binding kinetics of yHst2 D118N (blue diamonds) with data obtained in triplicate. (G) Dixon plot describing the nicotinamide inhibition of yHst2 D18N at varying concentrations of NAD+, 50 μM (blue diamonds), 100 μM (pink squares), 250 μM (yellow squares), 450 μM (cyan diamonds), 600 μM (purple circles), 800 μM (maroon circles), 1000 μM (blue minus signs), 1250 μM (green triangles), 1500 μM (cyan plus signs), 2000 μM (gray diamonds).
Figure 4. The overall structure of yHst2 I117F bound to carba-NAD+ and acetyl-lysine
(A) Simulated annealing omit density contoured at 1.0 sigma showing density for the protein (gray), carba-NAD+ (cyan), acetyllysine (magenta) and the mutated I117F residue (yellow). (B) The superposition of the nicotinamide bound yHst2/ADP-HPD/H4 and the yHst2 I117F/carba-NAD+/H4 structures. The yHst2 protein is shown in blue while the mutated I117F sidechain (yellow) and the nicotinamide molecule (red) are shown both in stick and modeled in terms of the van der Waals radius of each atom of the molecules.
Comment in
- Identification of a new nicotinamide binding site in a sirtuin: a reassessment.
Wolberger C. Wolberger C. Mol Cell. 2007 Dec 28;28(6):1102-3. doi: 10.1016/j.molcel.2007.12.006. Mol Cell. 2007. PMID: 18158906 No abstract available.
Similar articles
- Structural basis for nicotinamide cleavage and ADP-ribose transfer by NAD(+)-dependent Sir2 histone/protein deacetylases.
Zhao K, Harshaw R, Chai X, Marmorstein R. Zhao K, et al. Proc Natl Acad Sci U S A. 2004 Jun 8;101(23):8563-8. doi: 10.1073/pnas.0401057101. Epub 2004 May 18. Proc Natl Acad Sci U S A. 2004. PMID: 15150415 Free PMC article. - Sir2 regulation by nicotinamide results from switching between base exchange and deacetylation chemistry.
Sauve AA, Schramm VL. Sauve AA, et al. Biochemistry. 2003 Aug 12;42(31):9249-56. doi: 10.1021/bi034959l. Biochemistry. 2003. PMID: 12899610 - Mechanism of sirtuin inhibition by nicotinamide: altering the NAD(+) cosubstrate specificity of a Sir2 enzyme.
Avalos JL, Bever KM, Wolberger C. Avalos JL, et al. Mol Cell. 2005 Mar 18;17(6):855-68. doi: 10.1016/j.molcel.2005.02.022. Mol Cell. 2005. PMID: 15780941 - SIR2: the biochemical mechanism of NAD(+)-dependent protein deacetylation and ADP-ribosyl enzyme intermediates.
Sauve AA, Schramm VL. Sauve AA, et al. Curr Med Chem. 2004 Apr;11(7):807-26. doi: 10.2174/0929867043455675. Curr Med Chem. 2004. PMID: 15078167 Review. - Structure and chemistry of the Sir2 family of NAD+-dependent histone/protein deactylases.
Marmorstein R. Marmorstein R. Biochem Soc Trans. 2004 Dec;32(Pt 6):904-9. doi: 10.1042/BST0320904. Biochem Soc Trans. 2004. PMID: 15506920 Review.
Cited by
- Mechanistic studies on the effects of nicotinamide on megakaryocytic polyploidization and the roles of NAD+ levels and SIRT inhibition.
Giammona LM, Panuganti S, Kemper JM, Apostolidis PA, Lindsey S, Papoutsakis ET, Miller WM. Giammona LM, et al. Exp Hematol. 2009 Nov;37(11):1340-1352.e3. doi: 10.1016/j.exphem.2009.08.004. Epub 2009 Aug 26. Exp Hematol. 2009. PMID: 19715739 Free PMC article. - A molecular mechanism for direct sirtuin activation by resveratrol.
Gertz M, Nguyen GT, Fischer F, Suenkel B, Schlicker C, Fränzel B, Tomaschewski J, Aladini F, Becker C, Wolters D, Steegborn C. Gertz M, et al. PLoS One. 2012;7(11):e49761. doi: 10.1371/journal.pone.0049761. Epub 2012 Nov 21. PLoS One. 2012. PMID: 23185430 Free PMC article. - Structural basis for sirtuin activity and inhibition.
Yuan H, Marmorstein R. Yuan H, et al. J Biol Chem. 2012 Dec 14;287(51):42428-35. doi: 10.1074/jbc.R112.372300. Epub 2012 Oct 18. J Biol Chem. 2012. PMID: 23086949 Free PMC article. Review. - Structural analysis of trypanosomal sirtuin: an insight for selective drug design.
Kaur S, Shivange AV, Roy N. Kaur S, et al. Mol Divers. 2010 Feb;14(1):169-78. doi: 10.1007/s11030-009-9147-7. Epub 2009 Apr 29. Mol Divers. 2010. PMID: 19404761 - Deacylation Mechanism by SIRT2 Revealed in the 1'-SH-2'-O-Myristoyl Intermediate Structure.
Wang Y, Fung YME, Zhang W, He B, Chung MWH, Jin J, Hu J, Lin H, Hao Q. Wang Y, et al. Cell Chem Biol. 2017 Mar 16;24(3):339-345. doi: 10.1016/j.chembiol.2017.02.007. Epub 2017 Mar 9. Cell Chem Biol. 2017. PMID: 28286128 Free PMC article.
References
- Avalos JL, Bever KM, Wolberger C. Mechanism of sirtuin inhibition by nicotinamide: altering the NAD(+) cosubstrate specificity of a Sir2 enzyme. Mol Cell. 2005;17:855–868. - PubMed
- Avalos JL, Boeke JD, Wolberger C. Structural basis for the mechanism and regulation of Sir2 enzymes. Mol Cell. 2004;13:639–648. - PubMed
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–45107. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- DMR-0225180/GM/NIGMS NIH HHS/United States
- CA09171/CA/NCI NIH HHS/United States
- CA107107/CA/NCI NIH HHS/United States
- P41 RR001646/RR/NCRR NIH HHS/United States
- T32 CA009171/CA/NCI NIH HHS/United States
- RR-01646/RR/NCRR NIH HHS/United States
- R01 CA107107/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases