qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data - PubMed (original) (raw)
qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data
Jan Hellemans et al. Genome Biol. 2007.
Abstract
Although quantitative PCR (qPCR) is becoming the method of choice for expression profiling of selected genes, accurate and straightforward processing of the raw measurements remains a major hurdle. Here we outline advanced and universally applicable models for relative quantification and inter-run calibration with proper error propagation along the entire calculation track. These models and algorithms are implemented in qBase, a free program for the management and automated analysis of qPCR data.
Figures
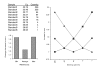
Figure 1
Effect of reference quantification cycle value on increase in error. Relative quantities were calculated for a simulated experiment with a five point four-fold dilution series using, respectively, the lowest Cq (squares), the average Cq (circles) or the highest Cq (triangles) as the reference quantification cycle value. Cq and quantity values are shown at the top left. The increase in the error on relative quantities for the different samples is shown at the top right, with the average increase depicted on the lower left graph.
Figure 2
Experimental setup. Experimental setup used to evaluate the effects of inter-run calibration. On the right side, a sample maximization approach is used to analyze 6 genes for 11 samples in 1.5 run. With gene maximization (left side), IRCs (S1, S2, S3) are required to allow comparison of S5-S7 (run 1) to S8-S11 (run 2 or 3), thus requiring two full runs. The IRCs in run 2 are measured on the same cDNA dilution whereas the IRCs in run 3 are measured on newly prepared cDNA from the same RNA.
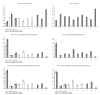
Figure 3
Experimental data comparing sample and gene maximization. The sample maximization approach (run 5) is compared to the gene maximization approach (runs 1 and 2 or 1 and 3). The difference between the IRCs is 0.77 for the Cq values, 72% for the NRQ values, and eliminated after inter-run calibration. Grey and white within the same display item indicates that data comes from different runs.
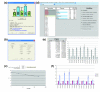
Figure 4
qBase. (a) qBase start up screen; (b) import wizard allowing selection of the format of the input file; (c) standard curve with a five point four-fold dilution series used to calculate the amplification efficiency; (d) qBase Analyzer main window with the workflow on the right and sample and gene list on the left - special sample types and reference genes are highlighted; (e) single gene histogram; (f) multi-gene histogram.
Figure 5
qBase calculation workflow.
Similar articles
- qpcR: an R package for sigmoidal model selection in quantitative real-time polymerase chain reaction analysis.
Ritz C, Spiess AN. Ritz C, et al. Bioinformatics. 2008 Jul 1;24(13):1549-51. doi: 10.1093/bioinformatics/btn227. Epub 2008 May 14. Bioinformatics. 2008. PMID: 18482995 - qPCR-DAMS: a database tool to analyze, manage, and store both relative and absolute quantitative real-time PCR data.
Jin N, He K, Liu L. Jin N, et al. Physiol Genomics. 2006 May 16;25(3):525-7. doi: 10.1152/physiolgenomics.00233.2005. Epub 2006 Feb 28. Physiol Genomics. 2006. PMID: 16507784 - Selection of reference genes for reverse transcription quantitative real-time PCR normalization in black rockfish (Sebastes schlegeli).
Liman M, Wenji W, Conghui L, Haiyang Y, Zhigang W, Xubo W, Jie Q, Quanqi Z. Liman M, et al. Mar Genomics. 2013 Sep;11:67-73. doi: 10.1016/j.margen.2013.08.002. Epub 2013 Sep 3. Mar Genomics. 2013. PMID: 24007945 - The real-time polymerase chain reaction.
Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonák J, Lind K, Sindelka R, Sjöback R, Sjögreen B, Strömbom L, Ståhlberg A, Zoric N. Kubista M, et al. Mol Aspects Med. 2006 Apr-Jun;27(2-3):95-125. doi: 10.1016/j.mam.2005.12.007. Epub 2006 Feb 3. Mol Aspects Med. 2006. PMID: 16460794 Review. - Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available.
Benes V, Castoldi M. Benes V, et al. Methods. 2010 Apr;50(4):244-9. doi: 10.1016/j.ymeth.2010.01.026. Epub 2010 Jan 28. Methods. 2010. PMID: 20109550 Review.
Cited by
- Influence of stress hormones on the auxin homeostasis in Brassica rapa seedlings.
Salopek-Sondi B, Šamec D, Mihaljević S, Smolko A, Pavlović I, Janković I, Ludwig-Müller J. Salopek-Sondi B, et al. Plant Cell Rep. 2013 Jul;32(7):1031-42. doi: 10.1007/s00299-013-1412-7. Epub 2013 Mar 19. Plant Cell Rep. 2013. PMID: 23508255 - Infection by Salmonella enterica Serovar Typhimurium DT104 Modulates Immune Responses, the Metabolome, and the Function of the Enteric Microbiota in Neonatal Broiler Chickens.
Bescucci DM, Montina T, Boras VF, Inglis GD. Bescucci DM, et al. Pathogens. 2022 Oct 29;11(11):1257. doi: 10.3390/pathogens11111257. Pathogens. 2022. PMID: 36365008 Free PMC article. - Validation of Tuba1a as appropriate internal control for normalization of gene expression analysis during mouse lung development.
Mehta A, Dobersch S, Dammann RH, Bellusci S, Ilinskaya ON, Braun T, Barreto G. Mehta A, et al. Int J Mol Sci. 2015 Feb 25;16(3):4492-511. doi: 10.3390/ijms16034492. Int J Mol Sci. 2015. PMID: 25723738 Free PMC article. - A reliable set of reference genes to normalize oxygen-dependent cytoglobin gene expression levels in melanoma.
De Backer J, Maric D, Bosman M, Dewilde S, Hoogewijs D. De Backer J, et al. Sci Rep. 2021 May 25;11(1):10879. doi: 10.1038/s41598-021-90284-6. Sci Rep. 2021. PMID: 34035373 Free PMC article. - Doenitin-1: A novel Kunitz family protein with versatile functions during feeding and reproduction of the tick Haemaphysalis doenitzi.
Lu J, Wang K, Gao Z, Zhang S, Li H, Shi Y, Song X, Liu J, Yu Z, Yang X. Lu J, et al. Front Vet Sci. 2022 Aug 10;9:872244. doi: 10.3389/fvets.2022.872244. eCollection 2022. Front Vet Sci. 2022. PMID: 36032296 Free PMC article.
References
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources