Structural basis for the actin-binding function of missing-in-metastasis - PubMed (original) (raw)
Structural basis for the actin-binding function of missing-in-metastasis
Sung Haeng Lee et al. Structure. 2007 Feb.
Abstract
The adaptor protein missing-in-metastasis (MIM) contains independent F- and G-actin binding domains, consisting, respectively, of an N-terminal 250 aa IRSp53/MIM homology domain (IMD) and a C-terminal WASP-homology domain 2 (WH2). We determined the crystal structures of MIM's IMD and that of its WH2 bound to actin. The IMD forms a dimer, with each subunit folded as an antiparallel three-helix bundle. This fold is related to that of the BAR domain. Like the BAR domain, the IMD has been implicated in membrane binding. Yet, comparison of the structures reveals that the membrane binding surfaces of the two domains have opposite curvatures, which may determine the type of curvature of the interacting membrane. The WH2 of MIM is longer than the prototypical WH2, interacting with all four subdomains of actin. We characterize a similar WH2 at the C terminus of IRSp53 and propose that in these two proteins WH2 performs a scaffolding function.
Figures
Figure 1
Crystal structure of the IMD of mouse MIM. (A) Schematic representation of MIM (yellow/blue, IMD; purple, middle regulatory/scaffolding region; red, WH2). (B) Ribbon representation of the structure of the IMD dimer (figure made with the program PyMOL,
). The two subunit of the dimer are colored blue and yellow. Helices 1 to 3 of each subunit are labeled H1, H2 and H3. Also shown is a sequence alignment corresponding to the conserved basic cluster at the symmetric ends of the IMD dimer (highlighted cyan in one of the subunits of the structure). In this alignment, red, blue, and green represent negatively charged, positively charged, and hydrophobic conserved amino acids, respectively. Accession numbers are: MIM_MOUSE, Q8R1S4; MIM_HUMAN, O43312; ABBA_HUMAN, Q765P7; IRSp53_HUMAN, Q9UQB8. Red arrows point to amino acids Leu 145 and Leu 147, which were mutated in this study. (C) Slice cut through the middle of the molecular surface of the IMD dimer revealing the interior cavity. (D) Close-up view of the “flap” loop between helices 3 and 4 that covers the “signature sequence” (Yamagishi et al., 2004) of the IMD, which is a charged and conserved sequence that is buried in the structure (red colored area of helix 3). (E) Superimposition of the structures of the IMDs of MIM and IRSp53. The two structures were superimposed based on the best overlapping central region (amino acids 26-68, 72-110 and 24-66, 69-107 of both chains of MIM and IRSp53, respectively). The view is as in part B and figure 3. This orientation highlights the differences between the A chains of the two proteins (blue). Although not well seen from this angle, similar differences occur between the B chains (yellow). Notice that helix 4 of the IMD of IRSp53 is missing in MIM.
Figure 2
Testing the actin binding and bundling activities of the IMD of MIM. (A) Purified IMD (left panel) and high-speed F-actin binding assay (right panel). 10 μM aliquots of IMD were ultracentrifuged at 400,000 X g, in the presence (+) and the absence (-) of 5 μM F-actin. Equal aliquots of supernatant (S) and pellet (P) were analyzed on a SDS-PAGE gel. (B) Quantification of the F-actin binding affinity of the IMD. 2 μM F-actin aliquots were incubated with increasing amounts of IMD (0, 1, 2, 4, 6, 8, 10, 15 and 20 μM) and analyzed on gel as in part 1A. Each point corresponds to the average densitometric reading of three independent experiments. The line represents the best fit of the data to the Michaelis-Menten equation. (C) High-speed co-sedimentation analysis of IMD mutants. 10 μM aliquots of each mutant were incubated with 5 μM F-actin and analyzed on gel (upper panel). The percentage of F-actin-bound IMD was calculated as the IMD fraction in pellet. Before each experiment the mutants were ultracentrifuged at 400,000 X g to eliminate potential aggregates. The lower panel illustrates the percentage of bound (empty bars) vs. unbound (black-filled bars) mutants from three independent experiments. (D) Low-speed analysis of the F-actin bundling activity of the IMD. 5 μM F-actin was centrifuged at 10,000 X g in the absence (-) and the presence (+) of 10 μM wild-type IMD, and the supernatant (S) and pellet (P) were analyzed on gel (upper panel). F-actin stays in the supernatant, indicating the lack of bundling activity. This result was confirmed by experiments carried out at increasing ratios IMD to F-actin (1, 2, 4, 6, 8, 10) (lower panel).
Figure 3
Structural and functional relationship between the IMD and BAR domains. (A) Electrostatic surface representation of the IMD dimer calculated with the program APBS (Baker et al., 2001) and displayed with the program PyMOL (
). Red and blue indicate negatively and positively charged regions, respectively (red, -6 kTe-1, blue +6 kTe-1). Note the positively charged and slightly convex surface, thought to mediate the interactions with membranes of the IMD (Suetsugu et al., 2006). (B) Similar electrostatic representation of the BAR domain of amphiphysin (Peter et al., 2004). The orientation is the same as in part A. Note that the shape of the positively charged membrane binding surface of the BAR domain is concave. (C) Superimposition of the structures of the IMD of MIM (blue, yellow) with that of the BAR domain of arfaptin complexed with Rac (Tarricone et al., 2001) (gray). The orientation is the same as in parts A and B. The two folds have different curvatures, but superimpose well in the middle section where the dimers overlap, suggesting that this region may also mediate the binding of Rac in MIM and IRSp53.
Figure 4
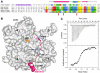
The WASP-homology domain 2 (WH2) of MIM and IRSp53. (A), Comparison of a classical WH2 (represented by WASP, Wiskott-Aldrich Syndrome Protein) with the WH2s of MIM, ABBA and IRSp53. Red, blue, green and yellow correspond to negatively charged, positively charged, hydrophobic and small (Thr, Val, Ser, Ala) conserved amino acids, respectively. The diagram above the sequences represents a secondary structure assignment based on the structure determined here (cylinder, α-helix; arrow, β-strand). Accession numbers are as in figure 1, and WASP_HUMAN, P42768. Red arrows point to non-canonical amino acids present in the WH2 of IRSp53. (B) Structure of the WH2 of MIM (red ribbon) bound to actin (gray surface). Numbers 1-4 indicate actin’s four subdomains. The side chains of some of the amino acids involved in interactions with actin are shown (green, hydrophobic; blue, positively charged). (C) Binding of the WH2 of IRSp53 to actin measured by ITC. The upper graph corresponds to the heat evolved upon repeated 10 μL injections of a 100 μM solution of the WH2 peptide into a 10 μM solution of actin in G-buffer. The lower graph shows the binding isotherm produced by integration of the heat for each injection. The line represents a nonlinear least squares fit to the data using a single-site binding model. The following thermodynamic parameters were determined from the fitting: dissociation constant Kd =0.28 ± 0.04 μM; molar enthalpy ΔH = -7.2 ± 0.1 kcal.mol-1; and stoichiometry n = 0.9.
Similar articles
- The structural basis of actin interaction with multiple WH2/beta-thymosin motif-containing proteins.
Aguda AH, Xue B, Irobi E, Préat T, Robinson RC. Aguda AH, et al. Structure. 2006 Mar;14(3):469-76. doi: 10.1016/j.str.2005.12.011. Structure. 2006. PMID: 16531231 - A novel actin bundling/filopodium-forming domain conserved in insulin receptor tyrosine kinase substrate p53 and missing in metastasis protein.
Yamagishi A, Masuda M, Ohki T, Onishi H, Mochizuki N. Yamagishi A, et al. J Biol Chem. 2004 Apr 9;279(15):14929-36. doi: 10.1074/jbc.M309408200. Epub 2004 Jan 29. J Biol Chem. 2004. PMID: 14752106 - Actin-bound structures of Wiskott-Aldrich syndrome protein (WASP)-homology domain 2 and the implications for filament assembly.
Chereau D, Kerff F, Graceffa P, Grabarek Z, Langsetmo K, Dominguez R. Chereau D, et al. Proc Natl Acad Sci U S A. 2005 Nov 15;102(46):16644-9. doi: 10.1073/pnas.0507021102. Epub 2005 Nov 7. Proc Natl Acad Sci U S A. 2005. PMID: 16275905 Free PMC article. - WH2 domain: a small, versatile adapter for actin monomers.
Paunola E, Mattila PK, Lappalainen P. Paunola E, et al. FEBS Lett. 2002 Feb 20;513(1):92-7. doi: 10.1016/s0014-5793(01)03242-2. FEBS Lett. 2002. PMID: 11911886 Review. - Control of actin assembly by the WH2 domains and their multifunctional tandem repeats in Spire and Cordon-Bleu.
Carlier MF, Husson C, Renault L, Didry D. Carlier MF, et al. Int Rev Cell Mol Biol. 2011;290:55-85. doi: 10.1016/B978-0-12-386037-8.00005-3. Int Rev Cell Mol Biol. 2011. PMID: 21875562 Review.
Cited by
- Downregulation of MTSS1 expression is an independent prognosticator in squamous cell carcinoma of the lung.
Kayser G, Csanadi A, Kakanou S, Prasse A, Kassem A, Stickeler E, Passlick B, Zur Hausen A. Kayser G, et al. Br J Cancer. 2015 Mar 3;112(5):866-73. doi: 10.1038/bjc.2015.2. Epub 2015 Jan 27. Br J Cancer. 2015. PMID: 25625275 Free PMC article. - Phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) specifically induces membrane penetration and deformation by Bin/amphiphysin/Rvs (BAR) domains.
Yoon Y, Zhang X, Cho W. Yoon Y, et al. J Biol Chem. 2012 Oct 5;287(41):34078-90. doi: 10.1074/jbc.M112.372789. Epub 2012 Aug 11. J Biol Chem. 2012. PMID: 22888025 Free PMC article. - Structural basis for the recruitment of profilin-actin complexes during filament elongation by Ena/VASP.
Ferron F, Rebowski G, Lee SH, Dominguez R. Ferron F, et al. EMBO J. 2007 Oct 31;26(21):4597-606. doi: 10.1038/sj.emboj.7601874. Epub 2007 Oct 4. EMBO J. 2007. PMID: 17914456 Free PMC article. - Structures of actin-bound Wiskott-Aldrich syndrome protein homology 2 (WH2) domains of Spire and the implication for filament nucleation.
Ducka AM, Joel P, Popowicz GM, Trybus KM, Schleicher M, Noegel AA, Huber R, Holak TA, Sitar T. Ducka AM, et al. Proc Natl Acad Sci U S A. 2010 Jun 29;107(26):11757-62. doi: 10.1073/pnas.1005347107. Epub 2010 Jun 10. Proc Natl Acad Sci U S A. 2010. PMID: 20538977 Free PMC article. - Dissecting BAR domain function in the yeast Amphiphysins Rvs161 and Rvs167 during endocytosis.
Youn JY, Friesen H, Kishimoto T, Henne WM, Kurat CF, Ye W, Ceccarelli DF, Sicheri F, Kohlwein SD, McMahon HT, Andrews BJ. Youn JY, et al. Mol Biol Cell. 2010 Sep 1;21(17):3054-69. doi: 10.1091/mbc.E10-03-0181. Epub 2010 Jul 7. Mol Biol Cell. 2010. PMID: 20610658 Free PMC article.
References
- Bompard G, Sharp SJ, Freiss G, Machesky LM. Involvement of Rac in actin cytoskeleton rearrangements induced by MIM-B. J Cell Sci. 2005;118:5393–5403. - PubMed
- Chereau D, Dominguez R. Understanding the role of the G-actin-binding domain of Ena/VASP in actin assembly. J Struct Biol. 2006 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases