Protein structural variation in computational models and crystallographic data - PubMed (original) (raw)
Protein structural variation in computational models and crystallographic data
Dmitry A Kondrashov et al. Structure. 2007 Feb.
Erratum in
- Structure. 2007 May;15(5):637
Abstract
Normal mode analysis offers an efficient way of modeling the conformational flexibility of protein structures. We use anisotropic displacement parameters from crystallography to test the quality of prediction of both the magnitude and directionality of conformational flexibility. Normal modes from four simple elastic network model potentials and from the CHARMM force field are calculated for a data set of 83 diverse, ultrahigh-resolution crystal structures. While all five potentials provide good predictions of the magnitude of flexibility, all-atom potentials have a clear edge at prediction of directionality, and the CHARMM potential has the highest prediction quality. The low-frequency modes from different potentials are similar, but those computed from the CHARMM potential show the greatest difference from the elastic network models. The comprehensive evaluation demonstrates the costs and benefits of using normal mode potentials of varying complexity.
Figures
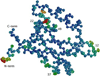
Figure 1
Example of a high-resolution protein structure (PDB ID 1R6J) showing anisotropic temperature factors for backbone atoms. The ellipsoids represent 90% probability volume of atomic position, with color varying from immobile (blue) to more mobile (red). Most of the backbone atoms are not mobile and isotropic, with a few loop residues (residue numbers labeled) showing clear directional preference in positional distribution.
Figure 2
Distributions of anisotropy parameters in structures refined with SHELX and Refmac software. The large difference is likely due to different default restraints on the anisotropic parameters in the two.
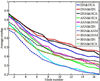
Figure 3
Overlap scores for individual normal modes from different potentials averaged over all 83 structures. Each curve is a comparison between a pair of potentials for the 17 lowest frequency modes. The solid curves compare different ENM-like potentials, while the dotted curves compare CHARMM-based BNM results with those from ENM potentials.
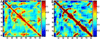
Figure 4
Correlation matrices generated from normal mode analyses of PDZ domain (PDB ID 1R6J). The plots show correlation between residues with indices shown on x and y axes, blue color indicating negative correlation and red signifying positive, with the range shown in the colorbars. Secondary structure elements are labeled in sequence order. A) Correlation from Anisotropic Network Model; B) from CHARMM-based Block Normal Modes. Note that the range in plot A is half that of B.
Comment in
- Normalizing normal mode analysis.
Chapman MS. Chapman MS. Structure. 2007 Feb;15(2):135-6. doi: 10.1016/j.str.2007.01.008. Structure. 2007. PMID: 17292830 No abstract available.
Similar articles
- Anisotropic fluctuations of amino acids in protein structures: insights from X-ray crystallography and elastic network models.
Eyal E, Chennubhotla C, Yang LW, Bahar I. Eyal E, et al. Bioinformatics. 2007 Jul 1;23(13):i175-84. doi: 10.1093/bioinformatics/btm186. Bioinformatics. 2007. PMID: 17646294 - Tensorial elastic network model for protein dynamics: integration of the anisotropic network model with bond-bending and twist elasticities.
Srivastava A, Halevi RB, Veksler A, Granek R. Srivastava A, et al. Proteins. 2012 Dec;80(12):2692-700. doi: 10.1002/prot.24153. Epub 2012 Aug 21. Proteins. 2012. PMID: 22847894 - Normal-mode refinement of anisotropic thermal parameters for potassium channel KcsA at 3.2 A crystallographic resolution.
Chen X, Poon BK, Dousis A, Wang Q, Ma J. Chen X, et al. Structure. 2007 Aug;15(8):955-62. doi: 10.1016/j.str.2007.06.012. Structure. 2007. PMID: 17698000 Free PMC article. - Atomic displacement parameters in structural biology.
Carugo O. Carugo O. Amino Acids. 2018 Jul;50(7):775-786. doi: 10.1007/s00726-018-2574-y. Epub 2018 May 11. Amino Acids. 2018. PMID: 29752562 Review. - Protein interactions and fluctuations in a proteomic network using an elastic network model.
Demirel MC, Keskin O. Demirel MC, et al. J Biomol Struct Dyn. 2005 Feb;22(4):381-6. doi: 10.1080/07391102.2005.10507010. J Biomol Struct Dyn. 2005. PMID: 15588102 Review.
Cited by
- A comparative analysis of the equilibrium dynamics of a designed protein inferred from NMR, X-ray, and computations.
Liu L, Koharudin LM, Gronenborn AM, Bahar I. Liu L, et al. Proteins. 2009 Dec;77(4):927-39. doi: 10.1002/prot.22518. Proteins. 2009. PMID: 19688820 Free PMC article. - Normal Mode Analysis as a Routine Part of a Structural Investigation.
Bauer JA, Pavlović J, Bauerová-Hlinková V. Bauer JA, et al. Molecules. 2019 Sep 10;24(18):3293. doi: 10.3390/molecules24183293. Molecules. 2019. PMID: 31510014 Free PMC article. Review. - Structural improvement of unliganded simian immunodeficiency virus gp120 core by normal-mode-based X-ray crystallographic refinement.
Chen X, Lu M, Poon BK, Wang Q, Ma J. Chen X, et al. Acta Crystallogr D Biol Crystallogr. 2009 Apr;65(Pt 4):339-47. doi: 10.1107/S0907444909003539. Epub 2009 Mar 19. Acta Crystallogr D Biol Crystallogr. 2009. PMID: 19307715 Free PMC article. - Molecular nonlinear dynamics and protein thermal uncertainty quantification.
Xia K, Wei GW. Xia K, et al. Chaos. 2014 Mar;24(1):013103. doi: 10.1063/1.4861202. Chaos. 2014. PMID: 24697365 Free PMC article. - All-atom contact model for understanding protein dynamics from crystallographic B-factors.
Li DW, Brüschweiler R. Li DW, et al. Biophys J. 2009 Apr 22;96(8):3074-81. doi: 10.1016/j.bpj.2009.01.011. Biophys J. 2009. PMID: 19383453 Free PMC article.
References
- Bahar I, Atilgan AR, Erman B. Direct evaluation of thermal fluctuations in proteins using a single-parameter harmonic potential. Fold Des. 1997;2:173–181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- 5T15LM007359/LM/NLM NIH HHS/United States
- T15 LM007359-01/LM/NLM NIH HHS/United States
- T15 LM007359/LM/NLM NIH HHS/United States
- T15 LM007359-04/LM/NLM NIH HHS/United States
- T15 LM007359-02/LM/NLM NIH HHS/United States
- T15 LM007359-03/LM/NLM NIH HHS/United States
- T15 LM007359-05/LM/NLM NIH HHS/United States
LinkOut - more resources
Full Text Sources