The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes - PubMed (original) (raw)
Comparative Study
. 2007 Apr;189(8):3228-36.
doi: 10.1128/JB.01726-06. Epub 2007 Feb 9.
Affiliations
- PMID: 17293413
- PMCID: PMC1855855
- DOI: 10.1128/JB.01726-06
Comparative Study
The genome sequence of avian pathogenic Escherichia coli strain O1:K1:H7 shares strong similarities with human extraintestinal pathogenic E. coli genomes
Timothy J Johnson et al. J Bacteriol. 2007 Apr.
Erratum in
- J Bacteriol. 2007 Jun;189(12):4554
Abstract
Escherichia coli strains that cause disease outside the intestine are known as extraintestinal pathogenic E. coli (ExPEC) and include human uropathogenic E. coli (UPEC) and avian pathogenic E. coli (APEC). Regardless of host of origin, ExPEC strains share many traits. It has been suggested that these commonalities may enable APEC to cause disease in humans. Here, we begin to test the hypothesis that certain APEC strains possess potential to cause human urinary tract infection through virulence genotyping of 1,000 APEC and UPEC strains, generation of the first complete genomic sequence of an APEC (APEC O1:K1:H7) strain, and comparison of this genome to all available human ExPEC genomic sequences. The genomes of APEC O1 and three human UPEC strains were found to be remarkably similar, with only 4.5% of APEC O1's genome not found in other sequenced ExPEC genomes. Also, use of multilocus sequence typing showed that some of the sequenced human ExPEC strains were more like APEC O1 than other human ExPEC strains. This work provides evidence that at least some human and avian ExPEC strains are highly similar to one another, and it supports the possibility that a food-borne link between some APEC and UPEC strains exists. Future studies are necessary to assess the ability of APEC to overcome the hurdles necessary for such a food-borne transmission, and epidemiological studies are required to confirm that such a phenomenon actually occurs.
Figures
FIG. 1.
Cluster analysis of 500 APEC and 500 UPEC strains for 39 ExPEC-associated traits. The leftmost portion of the figure is the dendrogram created from the average linkage cluster analysis on the presence of virulence factors. Just to the right of the dendrogram is column 1, which shows cluster membership based upon the dendrogram: red, cluster 1; cream, cluster 2; purple, cluster 3; orange, cluster 4; yellow, cluster 5; light blue, cluster 6; and green, cluster 7. Column 2 identifies an isolate as a UPEC strain (blue) or an APEC strain (green). Also, APEC O1 is identified in this column with a red bar. Columns 3 to 41 show the virulence genotype of each isolate tested. Each column in this group shows the results for a single gene. Black, gene is present; light green, gene is absent. Labels across the top of columns 3 to 41 show the APEC plasmid-linked genes (yellow) and the ExPEC chromosome-associated virulence genes (sky blue). ompTp, plasmid-encoded ompT; cvaB5, 5′ end of the cvaB gene; cvaB3, 3′ end of the cvaB gene; colM, cma; colB, cbi; papG1, papG allele 1; papG23, papG alleles 2 and 3. This method of analysis was first described by Rodriguez-Siek et al. (50) and has since been used in similar form elsewhere (8).
FIG. 2.
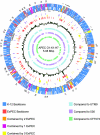
Map of the APEC O1 chromosome and comparison of APEC O1's genome to those of other ExPEC strains. The outer ring shows genomic islands, as numbers (1 to 43) corresponding to those in Table 5; the 2nd and 3rd rings are coding regions in forward and reverse orientation (blue, backbone ORFs also present in K-12; dark orange, ORFs absent from K-12 but present in all other sequenced ExPEC strains; light orange, ORFs absent from K-12 but present in APEC O1 and two other ExPEC strains; lavender, ORFs absent from K-12 but present in APEC O1 and one other ExPEC strain; yellow, ORFs present only in APEC O1); the 4th ring depicts the scale in base pairs; the 5th ring depicts tRNA genes in dark purple; the 6th ring depicts ORFs unique to APEC O1 in yellow; the 7th ring depicts ORFs common to all sequenced ExPEC strains in orange; the 8th ring shows sliding G+C content compared to the overall average of 50.6%; and the 9th, 10th, and 11th rings show genome alignments of APEC O1 with UTI89 (light blue), 536 (light purple), and CFT073 (light green), respectively.
FIG. 3.
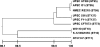
Results of MLST of fully sequenced E. coli genomes. The bottom line indicates percent similarity between strains. Sequence types (STs) are indicated to the right of the strain's name.
Similar articles
- Comparative genomic analysis shows that avian pathogenic Escherichia coli isolate IMT5155 (O2:K1:H5; ST complex 95, ST140) shares close relationship with ST95 APEC O1:K1 and human ExPEC O18:K1 strains.
Zhu Ge X, Jiang J, Pan Z, Hu L, Wang S, Wang H, Leung FC, Dai J, Fan H. Zhu Ge X, et al. PLoS One. 2014 Nov 14;9(11):e112048. doi: 10.1371/journal.pone.0112048. eCollection 2014. PLoS One. 2014. PMID: 25397580 Free PMC article. - Diversity and Population Overlap between Avian and Human Escherichia coli Belonging to Sequence Type 95.
Jørgensen SL, Stegger M, Kudirkiene E, Lilje B, Poulsen LL, Ronco T, Pires Dos Santos T, Kiil K, Bisgaard M, Pedersen K, Nolan LK, Price LB, Olsen RH, Andersen PS, Christensen H. Jørgensen SL, et al. mSphere. 2019 Jan 16;4(1):e00333-18. doi: 10.1128/mSphere.00333-18. mSphere. 2019. PMID: 30651401 Free PMC article. - Prevalence of avian-pathogenic Escherichia coli strain O1 genomic islands among extraintestinal and commensal E. coli isolates.
Johnson TJ, Wannemuehler Y, Kariyawasam S, Johnson JR, Logue CM, Nolan LK. Johnson TJ, et al. J Bacteriol. 2012 Jun;194(11):2846-53. doi: 10.1128/JB.06375-11. Epub 2012 Mar 30. J Bacteriol. 2012. PMID: 22467781 Free PMC article. - Avian pathogenic Escherichia coli (APEC).
Dho-Moulin M, Fairbrother JM. Dho-Moulin M, et al. Vet Res. 1999 Mar-Jun;30(2-3):299-316. Vet Res. 1999. PMID: 10367360 Review. - [Avian pathogenic Escherichia coli (APEC)].
Ewers C, Janssen T, Wieler LH. Ewers C, et al. Berl Munch Tierarztl Wochenschr. 2003 Sep-Oct;116(9-10):381-95. Berl Munch Tierarztl Wochenschr. 2003. PMID: 14526468 Review. German.
Cited by
- Identification and Functional Analysis of Novel Long Intergenic RNA in Chicken Macrophages Infected with Avian Pathogenic Escherichia coli.
Ma Y, Cao X, Sumayya, Lu Y, Han W, Lamont SJ, Sun H. Ma Y, et al. Microorganisms. 2024 Aug 6;12(8):1594. doi: 10.3390/microorganisms12081594. Microorganisms. 2024. PMID: 39203441 Free PMC article. - Molecular detection of avian pathogenic Escherichia coli (APEC) in broiler meat from retail meat shop.
Ranabhat G, Subedi D, Karki J, Paudel R, Luitel H, Bhattarai RK. Ranabhat G, et al. Heliyon. 2024 Aug 5;10(15):e35661. doi: 10.1016/j.heliyon.2024.e35661. eCollection 2024 Aug 15. Heliyon. 2024. PMID: 39170517 Free PMC article. - Tracing the Evolutionary Pathways of Serogroup O78 Avian Pathogenic Escherichia coli.
Ha EJ, Hong SM, Kim SJ, Ahn SM, Kim HW, Choi KS, Kwon HJ. Ha EJ, et al. Antibiotics (Basel). 2023 Dec 9;12(12):1714. doi: 10.3390/antibiotics12121714. Antibiotics (Basel). 2023. PMID: 38136748 Free PMC article. - Avian Pathogenic Escherichia coli (APEC) in Broiler Breeders: An Overview.
Joseph J, Zhang L, Adhikari P, Evans JD, Ramachandran R. Joseph J, et al. Pathogens. 2023 Oct 26;12(11):1280. doi: 10.3390/pathogens12111280. Pathogens. 2023. PMID: 38003745 Free PMC article. Review. - The role of AJB35136 and fdtA genes in biofilm formation by avian pathogenic Escherichia coli.
Khan MM, Ali A, Kolenda R, Olowe OA, Weinreich J, Li G, Schierack P. Khan MM, et al. BMC Vet Res. 2023 Aug 18;19(1):126. doi: 10.1186/s12917-023-03672-7. BMC Vet Res. 2023. PMID: 37596603 Free PMC article.
References
- Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. - PubMed
- Barnes, H. J., J. P. Vaillancourt, and W. B. Gross. 2003. Diseases of poultry, p. 631-652. Iowa State University Press, Ames, IA.
- Binnewies, T. T., Y. Motro, P. F. Hallin, O. Lund, D. Dunn, T. La, D. J. Hampson, M. Bellgard, T. M. Wassenaar, and D. W. Ussery. 2006. Ten years of bacterial genome sequencing: comparative-genomics-based discoveries. Funct. Integr. Genomics 6:165-185. - PubMed
- Blanco, M., J. E. Blanco, M. P. Alonso, and J. Blanco. 1994. Virulence factors and O groups of Escherichia coli strains isolated from cultures of blood specimens from urosepsis and non-urosepsis patients. Microbiologia 10:249-256. - PubMed
- Blanco, M., J. Blanco, J. E. Blanco, and J. Ramos. 1993. Enterotoxigenic, verotoxigenic, and necrotoxigenic Escherichia coli isolated from cattle in Spain. Am. J. Vet. Res. 54:1446-1451. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases