FMRP mediates mGluR5-dependent translation of amyloid precursor protein - PubMed (original) (raw)
FMRP mediates mGluR5-dependent translation of amyloid precursor protein
Cara J Westmark et al. PLoS Biol. 2007 Mar.
Abstract
Amyloid precursor protein (APP) facilitates synapse formation in the developing brain, while beta-amyloid (Abeta) accumulation, which is associated with Alzheimer disease, results in synaptic loss and impaired neurotransmission. Fragile X mental retardation protein (FMRP) is a cytoplasmic mRNA binding protein whose expression is lost in fragile X syndrome. Here we show that FMRP binds to the coding region of APP mRNA at a guanine-rich, G-quartet-like sequence. Stimulation of cortical synaptoneurosomes or primary neuronal cells with the metabotropic glutamate receptor agonist DHPG increased APP translation in wild-type but not fmr-1 knockout samples. APP mRNA coimmunoprecipitated with FMRP in resting synaptoneurosomes, but the interaction was lost shortly after DHPG treatment. Soluble Abeta40 or Abeta42 levels were significantly higher in multiple strains of fmr-1 knockout mice compared to wild-type controls. Our data indicate that postsynaptic FMRP binds to and regulates the translation of APP mRNA through metabotropic glutamate receptor activation and suggests a possible link between Alzheimer disease and fragile X syndrome.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
Figure 1. The Coding Region of APP mRNA Contains a Putative G-Quartet Sequence within a G-Rich Region Containing Several DWGG Repeats
(A) Alignment of the canonical G-quartet motif with the putative G-quartet sequence in human, mouse, and rat APP mRNAs. (B) Alignment of the G-rich region of the human, mouse, and rat APP genes. The predicted G-quartet sequence is located at position 825–846 of the mouse gene and is highlighted.
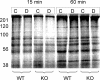
Figure 2. SNs Prepared from WT and fmr-1 KO Cortices Are Translationally Active
SDS-PAGE analysis of 35S-Met–labeled SNs without (C lanes) or with (D lanes) DHPG stimulation for times shown in minutes. The data are representative of multiple experiments: n = 6 (WT); n = 5 (KO).
Figure 3. mGluR Activation Increases APP Translation in SNs
(A) Immunoprecipitated, 35S-labeled APP (120-kDa band) from WT (15 min) and KO and WT (60 min) SNs analyzed by SDS-PAGE and (B) plotted as a percentage of APP synthesis; n = 3 repetitions. Asterisk indicates significant differences, with p = 0.008 between ±DHPG samples at 15 min and p = 0.016 between control at 15 min and DHPG at 60 min. For the control samples at 15 and 60 min, p = 0.056, and for the samples with or without DHPG at 60 min, p = 0.05. (C) Immunoprecipitated, 35S-labeled APP (120-kDa band) from WT SNs treated with DHPG, anisomycin, and MPEP, analyzed by SDS-PAGE, and (D) plotted as a percentage of APP synthesis; n = 3 repetitions (DHPG), n = 4 (anisomycin + DHPG and anisomycin), and n = 5 (MPEP + DHPG and MPEP).
Figure 4. Differential Regulation of APP Levels in WT and KO SNs
Western blots of WT (top panel) and KO (bottom panel) SN treated with or without DHPG (5, 10, and 20 min) and hybridized with anti-APP and anti–β-actin antibodies. The data are representative of three experiments, and quantitation with ImageQuant software demonstrates a 1.6–1.8-fold increase in APP between untreated and DHPG-stimulated WT SNs at all of the times tested.
Figure 5. DHPG Enhances APP Translation in WT but Not fmr-1 KO Neurons
(A) Immunofluorescent confocal images of WT (top) and KO (bottom) neuronal cells treated with or without DHPG (0, 10, and 20 min) and hybridized with anti-22C11 APP primary and anti-mouse rhodamine-conjugated secondary antibodies. The dashed yellow rectangles encompass segments of dendrites, which are enlarged and displayed below the photos. (B) Dendritic APP levels were quantitated with ImageJ software and plotted as a percentage of untreated WT samples. Asterisks indicate significant differences, with p < 0.001 between the pairs.
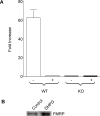
Figure 6. mGluR Activation Dislodges FMRP from APP mRNA
(A) APP mRNA was coimmunoprecipitated with FMRP from WT and KO SNs with or without DHPG treatment for 60 min, analyzed by RTqPCR, and plotted as the fold increase in APP mRNA. The data are the average of three experiments. (B) FMRP was immunoprecipitated from WT SNs with or without DHPG for 60 min and analyzed by Western blotting. The data are representative of two experiments.
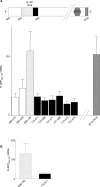
Figure 7. FMRP Binds to a G-Rich Sequence in APP mRNA
(A) Relative positions of the G-rich, predicted G-quartet and 29 base elements in nucleotides 446-2500 of APP (top). FMRP IPs digested with ribonuclease T1, analyzed by RTqPCR, and plotted as a percentage of APP mRNA699–796 (bottom). (B) FMRP IPs analyzed by the modified CLIP method and plotted as a percentage of APP699–796 mRNA.
Figure 8. Increased Aβ40 and Aβ42 Levels in fmr-1 KO Mice
(A) Soluble brain lysates from 1-y-old WT and fmr-1 KO mice (FVB strain) analyzed by ELISA and plotted as a percentage of soluble Aβ compared to WT controls. Student _t_-tests: p = 0.06 (Aβ40) and p = 0.001 (Aβ42). (B) GnHCl-soluble brain lysates from 1-y-old WT and fmr-1 KO mice (C57BL/6 strain) analyzed by ELISA and plotted as a percentage of GnHCl-soluble Aβ compared to WT controls. Student _t_-tests: p < 0.001 (Aβ40) and p = 0.39 (Aβ42).
Comment in
- Protein-RNA interaction links fragile X syndrome and Alzheimer disease.
Robinson R. Robinson R. PLoS Biol. 2007 Mar;5(3):e84. doi: 10.1371/journal.pbio.0050084. Epub 2007 Feb 13. PLoS Biol. 2007. PMID: 20076662 Free PMC article. No abstract available.
Similar articles
- The fragile X mental retardation protein is required for type-I metabotropic glutamate receptor-dependent translation of PSD-95.
Todd PK, Mack KJ, Malter JS. Todd PK, et al. Proc Natl Acad Sci U S A. 2003 Nov 25;100(24):14374-8. doi: 10.1073/pnas.2336265100. Epub 2003 Nov 12. Proc Natl Acad Sci U S A. 2003. PMID: 14614133 Free PMC article. - MPEP reduces seizure severity in Fmr-1 KO mice over expressing human Abeta.
Westmark CJ, Westmark PR, Malter JS. Westmark CJ, et al. Int J Clin Exp Pathol. 2009 Oct 10;3(1):56-68. Int J Clin Exp Pathol. 2009. PMID: 19918329 Free PMC article. - hnRNP C promotes APP translation by competing with FMRP for APP mRNA recruitment to P bodies.
Lee EK, Kim HH, Kuwano Y, Abdelmohsen K, Srikantan S, Subaran SS, Gleichmann M, Mughal MR, Martindale JL, Yang X, Worley PF, Mattson MP, Gorospe M. Lee EK, et al. Nat Struct Mol Biol. 2010 Jun;17(6):732-9. doi: 10.1038/nsmb.1815. Epub 2010 May 16. Nat Struct Mol Biol. 2010. PMID: 20473314 Free PMC article. - Autism, Alzheimer disease, and fragile X: APP, FMRP, and mGluR5 are molecular links.
Sokol DK, Maloney B, Long JM, Ray B, Lahiri DK. Sokol DK, et al. Neurology. 2011 Apr 12;76(15):1344-52. doi: 10.1212/WNL.0b013e3182166dc7. Neurology. 2011. PMID: 21482951 Free PMC article. Review. - Metabotropic glutamate receptors and fragile x mental retardation protein: partners in translational regulation at the synapse.
Ronesi JA, Huber KM. Ronesi JA, et al. Sci Signal. 2008 Feb 5;1(5):pe6. doi: 10.1126/stke.15pe6. Sci Signal. 2008. PMID: 18272470 Review.
Cited by
- Exploring cross-tissue DNA methylation patterns: blood-brain CpGs as potential neurodegenerative disease biomarkers.
Mendonça V, Soares-Lima SC, Moreira MAM. Mendonça V, et al. Commun Biol. 2024 Jul 26;7(1):904. doi: 10.1038/s42003-024-06591-x. Commun Biol. 2024. PMID: 39060467 Free PMC article. - RNA and neuronal function: the importance of post-transcriptional regulation.
Bhat VD, Jayaraj J, Babu K. Bhat VD, et al. Oxf Open Neurosci. 2022 Jul 7;1:kvac011. doi: 10.1093/oons/kvac011. eCollection 2022. Oxf Open Neurosci. 2022. PMID: 38596700 Free PMC article. Review. - Analysis of Nucleotide Variations in Human G-Quadruplex Forming Regions Associated with Disease States.
Neupane A, Chariker JH, Rouchka EC. Neupane A, et al. Genes (Basel). 2023 Nov 25;14(12):2125. doi: 10.3390/genes14122125. Genes (Basel). 2023. PMID: 38136947 Free PMC article. - On-Tissue Spatial Proteomics Integrating MALDI-MS Imaging with Shotgun Proteomics Reveals Soy Consumption-Induced Protein Changes in a Fragile X Syndrome Mouse Model.
Ma M, Yu Q, Delafield DG, Cui Y, Li Z, Li M, Wu W, Shi X, Westmark PR, Gutierrez A, Ma G, Gao A, Xu M, Xu W, Westmark CJ, Li L. Ma M, et al. ACS Chem Neurosci. 2024 Jan 3;15(1):119-133. doi: 10.1021/acschemneuro.3c00497. Epub 2023 Dec 18. ACS Chem Neurosci. 2024. PMID: 38109073 Free PMC article. - Ketogenic Diet Affects Sleep Architecture in C57BL/6J Wild Type and Fragile X Mice.
Westmark PR, Gholston AK, Swietlik TJ, Maganti RK, Westmark CJ. Westmark PR, et al. Int J Mol Sci. 2023 Sep 22;24(19):14460. doi: 10.3390/ijms241914460. Int J Mol Sci. 2023. PMID: 37833907 Free PMC article.
References
- Akaaboune M, Allinquant B, Farza H, Roy K, Magoul R, et al. Developmental regulation of amyloid precursor protein at the neuromuscular junction in mouse skeletal muscle. Mol Cell Neurosci. 2000;15:355–367. - PubMed
- Kamenetz F, Tomita T, Hsieh H, Seabrook G, Borchelt D, et al. APP processing and synaptic function. Neuron. 2003;37:925–937. - PubMed
- Hagerman RJ, Hagerman PJ. In: Physical and behavioral phenotype. Hagerman RJ, Cronister A, editors. Baltimore: John Hopkins University Press; 2002. pp. 3–109.
- Rudelli RD, Brown WT, Wisniewski K, Jenkins EC, Laure-Kamionowska M, et al. Adult fragile X syndrome. Clinico-neuropathologic findings. Acta Neuropathol (Berl) 1985;67:289–295. - PubMed
- Wisniewski KE, Segan SM, Miezejeski CM, Sersen EA, Rudelli RD. The Fra(X) syndrome: Neurological, electrophysiological, and neuropathological abnormalities. Am J Med Genet. 1991;38:476–480. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous