Shugoshin 2 regulates localization of the chromosomal passenger proteins in fission yeast mitosis - PubMed (original) (raw)
Shugoshin 2 regulates localization of the chromosomal passenger proteins in fission yeast mitosis
Vincent Vanoosthuyse et al. Mol Biol Cell. 2007 May.
Abstract
Fission yeast has two members of the Shugoshin family, Sgo1 and Sgo2. Although Sgo1 has clearly been established as a protector of centromere cohesion in meiosis I, the roles of Sgo2 remain elusive. Here we show that Sgo2 is required to ensure proper chromosome biorientation upon recovery from a prolonged spindle checkpoint arrest. Consistent with this, Sgo2 is essential for maintaining the Passenger proteins on centromeres upon checkpoint activation. Interestingly, lack of Sgo2 has a more penetrant effect on the localization of Survivin than on the two other Passenger proteins INCENP and Aurora B, and the Survivin-INCENP complex but not the INCENP-Aurora B complex is destabilized in the absence of Sgo2. Finally we show that the conserved C-terminus of Sgo2 is crucial to maintain Sgo2 and Passenger proteins localization on centromeres upon prolonged checkpoint activation. Taken together, our results demonstrate that Sgo2 is important for chromosome biorientation and that it controls docking of the Passenger proteins on chromosomes in early mitotic cells.
Figures
Figure 1.
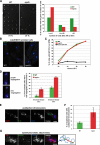
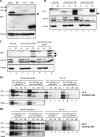
Sgo2 is required for proper chromosome biorientation after a prolonged spindle checkpoint arrest. (A) Cells lacking Sgo2 die after a _nda3KM311_-dependent arrest. Cells were shifted to 18°C for 8 h, and then individual cells were isolated on plates (rich medium) and left at 30°C for 3 d. The numbers under the pictures indicate the corresponding cell viability for that particular experiment (% of isolated cells forming colonies). The right panel quantifies the number of cells per individual colony after 24 h at 30°C. (B) nda3KM311 and _sgo2_Δ nda3KM311 cells arrested at 18°C for 8 h recruit a similar amount of the spindle checkpoint kinase Bub1 on kinetochores. Scale bar, 2 μm. (C) nda3KM311 GFPplo1, sgo2Δ nda3KM311 GFP-plo1, and bub1_Δ_20-160 nda3-KM311 GFPplo1 cells were incubated at 18°C for 8 h, and the number of metaphase cells was scored, using the strong enrichment of GFPplo1 on SPB after SPB duplication (Mulvihill et al., 1999). Cut12-CFP was used as a SPB marker for confirmation (not shown). Wild-type and _sgo2_Δ cells accumulated metaphase cells with similar kinetics after the shift at 18°C. On the contrary, the checkpoint-deficient allele bub1_Δ_28-160 (Vanoosthuyse et al., 2004) did not show any significant accumulation of metaphase cells. Furthermore in all backgrounds except bub1_Δ_28-160 chromosomes remained hyper-condensed and Cdc13/cyclinB and Cdc2 levels were maintained on SPBs (not shown). (D) nda3KM311 and _sgo2_Δ nda3KM311 cells were shifted at 18°C for either 6 or 8 h to arrest them in early mitosis. They were then released at 30 or 36°C, respectively, to allow anaphase onset, and the segregation of chromosome II was monitored in binucleate cells using GFP-tagged chromosomes (Ding et al., 2004). A minimum of 800 binucleate cells has been counted in total after a minimum of three independent experiments. (E) Example of anaphase with unattached chromosomes observed in the absence of Sgo2 upon release from an _nda3KM311_-dependent arrest. Cells were shifted at 18°C for 8 h, released at 36°C for 10 min, and then fixed with paraformaldehyde and processed for immunofluorescence with antibodies recognizing tubulin (tub) or the kinetochore component CNP1. The arrow indicates the two pairs of unattached sister-centromeres. Scale bar, 2 μm. (F) Quantification of the previous. (G) Example of monotelic/syntelic attachment. See diagram on the right of the picture for explanation. In that particular example, chromosome 3 (labeled “3” on the diagram) is easily recognizable by the two rDNA protrusions it carries (arrows). The two unseparated kinetochores of chromosome 3 are found at the left pole. Consistent with this, the four other kinetochores (chromosomes 1 and 2) are found on the spindle, two at the poles and two lagging. Finally, the fact that the 2 rDNA protrusions of chromosome 3 are facing away from the spindle is the final argument to conclude that chromosome 3 has not segregated. Compare to E for a normal anaphase segregation of chromosome 3 with its rDNA protrusions segregating in between the two poles. Scale bar, 2 μm.
Figure 2.
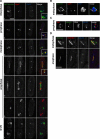
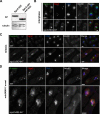
Sgo2 and the Passenger proteins colocalize in early mitosis before anaphase. (A) Sgo2-mCherry, GFP-Bir1/Survivin, and the SPB component Cut12 were observed concomitantly. To do so, we used a strain expressing 1) Sgo2 as a fusion with the monomeric red fluorescent protein mCherry (Shaner et al., 2004) (Sgo2-mCH), 2) the Spindle Pole Body (SPB) component Cut12 as a fusion with CFP (Cut12-CFP) and 3) Bir1/Survivin fused to GFP (GFP-Bir1/Survivin). All protein fusions were integrated at the endogenous locus and are functional. Cells 1–7 were selected from a cycling population and are representative of each individual stage of the cell cycle. Blowups of panels 1–4 are shown in the merge channel. Note that in early anaphase (panel 4) Survivin and Sgo2 showed greatly reduced colocalization at the poles. Survivin localization was consistent with a localization at the SPBs, whereas Sgo2 most likely localized on centromeres. (B) Bir1/Survivin localizes on SPBs every mitosis, as shown by its colocalization with the SPB component Sad1. (C) Bir1/Survivin localizes on telomeres every mitosis, as shown by its colocalization with the telomere component Taz1. (D) GFP-Bir1/Survivin and Pic1/INCENP-mCherry colocalize at all stages of the cell cycle. Examples of metaphase and anaphase are shown. Ark1/Aurora B displayed the same localization pattern (not shown and Morishita et al., 2001). Scale bar, 3 μm in all panels.
Figure 3.
Lack of Sgo2 affects the localization of the Passenger proteins. Scale bar, 2 μm in all panels. (A) Bir1/Survivin and Sgo2 localize only on centromeres in _nda3KM311_-arrested cells. (B) Bir1/Survivin and Pic1/INCENP colocalize on centromeres in _nda3KM311_-arrested cells. (C) In the absence of Sgo2, Bir1/Survivin failed to localize on centromeres in _nda3KM311_-arrested cells and instead colocalized with the SPB marker Cut12 (D). Note that in both C and D Bir1/Survivin was also found in the nucleus in the absence but not in the presence of Sgo2. (E) nda3KM311 cells expressing Ark1/Aurora B-GFP and the kinetochore marker Ndc80-CFP were arrested at 18°C in the presence or the absence of the _Sgo2_+ gene. Only cells where the three kinetochore pairs were clearly individualized were examined (3D analysis and deconvolution on 55 cells). In these cells, it was straightforward to determine whether Ark1/Aurora B localized on centromeres or on SPBs. In the absence of _Sgo2_+ (_sgo2_Δ), Ark1/Aurora B was clearly mislocalized on SPBs in 80% of the cells but remained on centromeres at a drastically reduced level in 20% of the cells. The pictures _Sgo2_+ and _sgo2_Δ were taken in exactly the same conditions. (F) In the absence of Sgo2, none of the Passenger proteins localize on telomeres in early mitosis. A representative metaphase cell expressing Pic1/INCENP-GFP and the kinetochore marker Ndc80-CFP is shown in the presence (sgo2+) or in the absence of Sgo2 (sgo2Δ). Note that no signal outside the spindle axis (arrow) is observed in the absence of Sgo2. (G) Centromere localization of the Passenger proteins in the absence of Sgo2 in a normal mitosis. Metaphase sgo2Δ cells of a cycling population were analyzed by 3D capture and deconvolution. Three types of cells were identified (representative pictures of cells expressing Pic1/INCENP-GFP and Ndc80-CFP are placed on the right of the histogram). In the first type, the Passenger protein localized on centromeres; in the second type, it localized on SPBs, and in the third type, it localized prematurely on the spindle. The histograms show the percentage of cells found in each category for each of the three Passenger proteins. (H) In metaphase of cells lacking Sgo2, centromeres recruit only half the wild-type amount of Ark1/Aurora BGFP (see text for details). (I) Ark1/Aurora B-GFP is stable in the absence of Sgo2. Whole cell extracts of _Sgo2_+ or sgo2Δ cells arrested using the nda3KM311 mutation were analyzed by Western blot. Tubulin provided the loading control.
Figure 4.
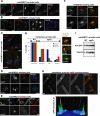
Sgo2 localization on centromeres is abolished by the Bir1/Survivin mutation bir1.46. (A) bir1.46 cells expressing Sgo2-GFP were grown overnight at 25°C, shifted or not at the restrictive temperature of 36°C, and then processed for immunolocalization of Sgo2 (α-GFP antibody) and the kinetochore marker Cnp1 (α-CNP1 antibody, kind gift of Prof. R. Allshire). In metaphase cells of bir1.46 cells, Sgo2 did not concentrate on centromeres and instead localized in the whole nucleus at both temperatures. Scale bar, 2 μm (B) The bir1.46 mutation did not affect the levels of Sgo2 in the cell, as shown by Western blot with our anti-Sgo2 antibody. Tubulin was used as a loading control.
Figure 5.
Lack of Sgo2 specifically destabilized the complex between Bir1/Survivin and Pic1/INCENP in checkpoint-arrested cells. (A) Ark1/Aurora B, Pic1/INCENP, Bir1/Survivin, and Sgo2 were all tagged with a SZZ tag. Whole cell extracts of each tagged strains were processed by Western blot, and the levels of expression of each construct were assessed with the Peroxidase anti-Peroxidase (PAP) antibody that recognizes the ZZ tag. Tubulin provided a loading control. The asterisk highlights a protein recognized aspecifically by the PAP antibody. (B) Ark1/Aurora B coimmunoprecipitates with Pic1/INCENP, but not with Bir1/Survivin. Bir1/Survivin-SZZ or Pic1/ INCENP-SZZ was pulled down on IgG Sepharose beads from _nda3-KM3111_-arrested cells. The complexes immunoprecipitated were assessed by Western blot for the presence of Ark1/Aurora B tagged with the Pk epitope (Ark1/Aurora B-Pk). A little Ark1/Aurora B-Pk sticks to the IgG beads even in the absence of any SZZ-tagged protein in the extract (see lane 2). The amount of Ark1/Aurora B-Pk pulled down by the IgG beads is dramatically increased in the presence of Pic1/INCENP-SZZ but not in the presence of Bir1/Survivin-SZZ (compare lanes 5 and 8). (C) Lack of Sgo2 does not affect the stability of the Pic1/INCENP-Ark1/Aurora B complex. Pic1/ INCENP-SZZ was pulled down on IgG Sepharose beads from _nda3-KM3111_-arrested cells. The complexes immunoprecipitated were assessed by Western blot for the presence of Ark1/Aurora B tagged with the Pk epitope (Ark1/Aurora B-Pk). There is consistently no significant difference in the amount of Ark1/Aurora B-Pk pulled down by Pic1/INCENP in the presence or absence of Sgo2 (compare lanes 5 and 8). The experiments described in B and C have been repeated at least four times. For B–D: In, input; Un, unbound; PD, pulldown. The arrows indicate Pic1/INCENP degradation products that occurred during the 40-min immunoprecipitation. These degradation products are not found in the total extracts (IN) or in extracts prepared from methanol-fixed cells (not shown). (D) Lack of Sgo2 reduces the interaction between Pic1/INCENP and Bir1/Survivin in checkpoint-arrested cells (D1) but not in interphase cells (D2). Pic1/INCENP-SZZ was pulled down as in B with the exception that IgG dynabeads were used instead of IgG Sepharose beads. The complexes immunoprecipitated were assessed for the presence of Bir1/Survivin tagged with the myc epitope (Bir1/Survivin-myc) using a polyclonal rabbit anti-myc antibody (A14, Santa Cruz Biotechnology, Santa Cruz, CA). The same antibody also recognized the TAPTAG on Pic1/INCENP. (D1) Extracts were prepared from cells arrested in mitosis at 18°C using the tubulin mutation nda3KM311. Serial dilutions of both extracts and immunoprecipitated complexes were loaded on the gel. Compare particularly lanes 2 and 6. We consistently observed a 2–5-fold reduction in the amount of Bir1/Survivin recovered in the Pic1/INCENP immunoprecipitated complexes in the absence of Sgo2. This experiment has been carried out 10 times and a significant reduction was observed every time. In this particular example, we observed a threefold reduction in the level of Bir1/Survivin recovered when lanes 2 and 6 were quantified. Despite a slight underloading of the _sgo2_Δ extracts, we chose to show this particular example because the amount of Pic1/INCENP recovered in the immunoprecipitated complexes was very similar between the Sgo2+ and _sgo2_Δ extracts. Lack of Sgo2 did not consistently affect the stability of either Pic1/INCENP or Bir1/Survivin (see D2 and Supplementary Figure 6). (D2) The same experiment as in D1 was performed with protein extracts made from cycling cells (30°C). Here, lack of Sgo2 had no effect on the amount of Bir1/Survivin recovered after Pic1/INCENP immunoprecipitation. In D1 and D2, the checkpoint component Mad1 was used as a loading control. In D2, extracts were also probed with an antibody directed against Sgo2 to confirm that the extracts were Sgo2+ or _sgo2_Δ.
Figure 6.
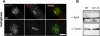
The conserved C-terminus of Sgo2 is required for Sgo2 localization on centromeres upon checkpoint arrest. Scale bar, 2 μm in all panels. (A) The truncated protein Sgo2Δ563-647 is stable. Whole cell extracts of the indicated strains were analyzed by Western blot. Tubulin provided a loading control. (B) The truncated protein Sgo2Δ563-647 was tagged with mCherry, and its localization pattern was analyzed in cycling cells concomitantly with GFP-Bir1/Survivin and the kinetochore marker Ndc80CFP. Strikingly the truncated protein was not enriched on telomeres in interphase but instead was found over the chromatin. (C) In mitosis of a cycling population, neither GFP-Bir1/Survivin nor Sgo2Δ563-647 localized on telomeres. However, they still localized on centromeres. Interestingly in anaphase, the truncated protein Sgo2Δ563-647 localized on the spindle (arrow). This is consistent with the idea that the conserved N-terminus of the Shugoshin family is a microtubule-binding domain (Salic et al., 2004). (D) In _nda3KM311_-arrested cells Sgo2Δ563-647 localized exclusively in the nucleus and no more than two GFP-Bir1/Survivin foci were detected, indicating that GFP-Bir1/Survivin localized on SPBs (see Figure 2).
Figure 7.
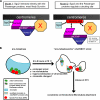
(A) Models for Sgo2 function. In the first model, Sgo2 forms a complex with Survivin. This complex, together with an additional factor X (see text for details) are crucial for Passenger proteins targeting to centromeres. The arrows indicate that Survivin and Sgo2 are interdependent for their localization on centromeres. In the second model, Sgo2 does not interact directly with the Passenger proteins but instead regulates their docking site. The stable interaction between the Passenger proteins and their docking site is in turn crucial for Sgo2 localization. (B) Diagram highlighting the wide range of changes in _nda3KM311_-arrested cells compared with normal prometaphase cells. At the restrictive temperature of 18°C, chromosomes hypercondense, the nuclear envelope loses its circular shape, and duplicated SPBs move away from each other, maintaining or not connections with one or more kinetochores. Furthermore, we speculate that chromatin changes (highlighted by a yellow line around the chromosomes) could enhance the association of the Passenger proteins with centromeres (see Discussion). On release at 36°C, the nuclear geometry generated in the arrest constrains kinetochore-microtubules attachments.
Similar articles
- Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres.
Kawashima SA, Tsukahara T, Langegger M, Hauf S, Kitajima TS, Watanabe Y. Kawashima SA, et al. Genes Dev. 2007 Feb 15;21(4):420-35. doi: 10.1101/gad.1497307. Genes Dev. 2007. PMID: 17322402 Free PMC article. - The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis.
Kitajima TS, Kawashima SA, Watanabe Y. Kitajima TS, et al. Nature. 2004 Feb 5;427(6974):510-7. doi: 10.1038/nature02312. Epub 2004 Jan 18. Nature. 2004. PMID: 14730319 - Control of Shugoshin function during fission-yeast meiosis.
Vaur S, Cubizolles F, Plane G, Genier S, Rabitsch PK, Gregan J, Nasmyth K, Vanoosthuyse V, Hardwick KG, Javerzat JP. Vaur S, et al. Curr Biol. 2005 Dec 20;15(24):2263-70. doi: 10.1016/j.cub.2005.11.034. Curr Biol. 2005. PMID: 16360688 - Role of chromosomal passenger complex in chromosome segregation and cytokinesis.
Terada Y. Terada Y. Cell Struct Funct. 2001 Dec;26(6):653-7. doi: 10.1247/csf.26.653. Cell Struct Funct. 2001. PMID: 11942622 Review. - Chromosomal passengers and the (aurora) ABCs of mitosis.
Adams RR, Carmena M, Earnshaw WC. Adams RR, et al. Trends Cell Biol. 2001 Feb;11(2):49-54. doi: 10.1016/s0962-8924(00)01880-8. Trends Cell Biol. 2001. PMID: 11166196 Review.
Cited by
- Sequential assembly of centromeric proteins in male mouse meiosis.
Parra MT, Gómez R, Viera A, Llano E, Pendás AM, Rufas JS, Suja JA. Parra MT, et al. PLoS Genet. 2009 Mar;5(3):e1000417. doi: 10.1371/journal.pgen.1000417. Epub 2009 Mar 13. PLoS Genet. 2009. PMID: 19283064 Free PMC article. - Schizosaccharomyces pombe Bub3 is dispensable for mitotic arrest following perturbed spindle formation.
Tange Y, Niwa O. Tange Y, et al. Genetics. 2008 Jun;179(2):785-92. doi: 10.1534/genetics.107.081695. Epub 2008 May 27. Genetics. 2008. PMID: 18505884 Free PMC article. - Malonylation of histone H2A at lysine 119 inhibits Bub1-dependent H2A phosphorylation and chromosomal localization of shugoshin proteins.
Ishiguro T, Tanabe K, Kobayashi Y, Mizumoto S, Kanai M, Kawashima SA. Ishiguro T, et al. Sci Rep. 2018 May 16;8(1):7671. doi: 10.1038/s41598-018-26114-z. Sci Rep. 2018. PMID: 29769606 Free PMC article. - Xenopus Shugoshin 2 regulates the spindle assembly pathway mediated by the chromosomal passenger complex.
Rivera T, Ghenoiu C, Rodríguez-Corsino M, Mochida S, Funabiki H, Losada A. Rivera T, et al. EMBO J. 2012 Mar 21;31(6):1467-79. doi: 10.1038/emboj.2012.4. Epub 2012 Jan 24. EMBO J. 2012. PMID: 22274615 Free PMC article. - Int6 and Moe1 interact with Cdc48 to regulate ERAD and proper chromosome segregation.
Otero JH, Suo J, Gordon C, Chang EC. Otero JH, et al. Cell Cycle. 2010 Jan 1;9(1):147-61. doi: 10.4161/cc.9.1.10312. Epub 2010 Jan 9. Cell Cycle. 2010. PMID: 20016281 Free PMC article.
References
- Allshire R. C., Nimmo E. R., Ekwall K., Javerzat J. P., Cranston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. - PubMed
- Delacour-Larose M., Molla A., Skoufias D. A., Margolis R. L., Dimitrov S. Distinct dynamics of Aurora B and Survivin during mitosis. Cell Cycle. 2004;3:1418–1426. - PubMed
- Ding D. Q., Yamamoto A., Haraguchi T., Hiraoka Y. Dynamics of homologous chromosome pairing during meiotic prophase in fission yeast. Dev. Cell. 2004;6:329–341. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases