An unmethylated 3' promoter-proximal region is required for efficient transcription initiation - PubMed (original) (raw)
An unmethylated 3' promoter-proximal region is required for efficient transcription initiation
Ruth Appanah et al. PLoS Genet. 2007.
Abstract
The promoter regions of approximately 40% of genes in the human genome are embedded in CpG islands, CpG-rich regions that frequently extend on the order of one kb 3' of the transcription start site (TSS) region. CpGs 3' of the TSS of actively transcribed CpG island promoters typically remain methylation-free, indicating that maintaining promoter-proximal CpGs in an unmethylated state may be important for efficient transcription. Here we utilize recombinase-mediated cassette exchange to introduce a Moloney Murine Leukemia Virus (MoMuLV)-based reporter, in vitro methylated 1 kb downstream of the TSS, into a defined genomic site. In a subset of clones, methylation spreads to within approximately 320 bp of the TSS, yielding a dramatic decrease in transcript level, even though the promoter/TSS region remains unmethylated. Chromatin immunoprecipitation analyses reveal that such promoter-proximal methylation results in loss of RNA polymerase II and TATA-box-binding protein (TBP) binding in the promoter region, suggesting that repression occurs at the level of transcription initiation. While DNA methylation-dependent trimethylation of H3 lysine (K)9 is confined to the intragenic methylated region, the promoter and downstream regions are hypo-acetylated on H3K9/K14. Furthermore, DNase I hypersensitivity and methylase-based single promoter analysis (M-SPA) experiments reveal that a nucleosome is positioned over the unmethylated TATA-box in these clones, indicating that dense DNA methylation downstream of the promoter region is sufficient to alter the chromatin structure of an unmethylated promoter. Based on these observations, we propose that a DNA methylation-free region extending several hundred bases downstream of the TSS may be a prerequisite for efficient transcription initiation. This model provides a biochemical explanation for the typical positioning of TSSs well upstream of the 3' end of the CpG islands in which they are embedded.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
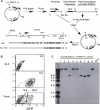
Figure 1. Generation and Targeting of the Patch-Methylated Reporter Cassette
(A) The construct L1-LTRGFP-1L was digested with HindIII (H) and BglII (Bgl), generating two fragments, one containing the LTR and ∼1 kb of the transcription unit and the other containing the 3′ end of the transcription unit, which includes the GFP gene. The latter fragment was methylated in vitro and ligated to the unmethylated promoter fragment, regenerating the original plasmid. RL5 MEL cells were cotransfected with this “patch”-methylated plasmid or an unmethylated control in combination with the Cre-expression plasmid CMV-Cre. Successful recombination between the loxP sites flanking the LTRGFP transgene and the pre-existing HyTK cassette yielded a targeted L1-LTRGFP-1L cassette initially methylated at the CpG sites as shown (L1 and 1L, loxP sites; SD, splice donor; SA, splice acceptor; pA, SV40 polyA). (B) FACS analyses of the RL5 parent line and ganciclovir resistant “pools” harboring the unmethylated (−) or patch-methylated L1-LTRGFP-1L cassette are shown as 5% probability plots. Median fluorescence values for each are shown, with the GFP+ patch-methylated population divided into two populations via gating, as shown. (C) Southern analysis of genomic DNA isolated from clones harboring the unmethylated L1-LTRGFP-1L cassette was conducted using a GFP probe. With the exception of clone 7, all clones harbor the cassette integrated in the RL5 site in one of two possible orientations.
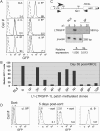
Figure 2. The Patch-Methylated L1-LTRGFP-1L Cassette is Expressed at a Low Level in a Subset of Clones
(A) Orientation-matched, unmethylated (−) and patch-methylated (P) clones were analyzed for GFP expression by flow cytometry along with the RL5 parent cell line (dashed line), which is under-laid with each clone for reference. Median fluorescence values are displayed in the lower right hand corner of each histogram. (B) The median GFP fluorescence values of RL5, clone 6− and 10 patch-methylated clones analyzed at day 38 post-RMCE are shown. Clone 9P (marked with an asterisk) was chosen for further study. (C) Quantitative duplex RT-PCR analysis of total RNA isolated from a representative low-expressing patch-methylated clone (9P), the unmethylated control clone 6−, and the RL5 parent line was conducted using primers (horizontal arrows) specific for the spliced transgenic mRNA and the endogenous β-actin gene. Relative expression level was determined by normalizing the transgene-specific amplification product to the β-actin gene product. (D) Two sets of electronic gates were applied to sort relatively low- and high-expressing cells from clone 9P. A total of 104 viable cells of each subpopulation were sorted and cultured for five days prior to reanalysis by flow cytometry. The median fluorescence values for each sorted pool are shown in the lower right hand corner of each histogram, along with that of the RL5 parent line, and clone 6− for reference.
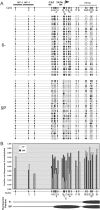
Figure 3. The Low-Expressing Patch-Methylated Clones Show Spreading of Methylation towards the Promoter Region
(A) A map of the integrated L1-LTRGFP-1L cassette showing the relevant restriction sites, the probe used for Southern analysis (horizontal bar), the primer pairs used for bisulphite sequencing, and a CpG density map. (B) Southern analysis of genomic DNA isolated from patch-methylated clones digested with BamHI alone or in combination with the methylation-sensitive restriction enzyme HpaII. The double digest reveals bands between ∼1660 bps and ∼530 bps, corresponding to the first HpaII site upstream of the methylated region (shown as an asterisk in [A]), or the fragment size predicted for complete digestion of the cassette, respectively. (C) The methylation status of representative high- (5P) and low- (9P) expressing patch-methylated clones and an unmethylated (6−) control was analyzed in greater detail by bisulphite sequencing, using primers specific for the LTR (I), intron (II), and downstream/methylated regions (IV). The presence of methylation at specific CpG sites is shown (black ovals) for each molecule sequenced (horizontal lines). Empty ovals indicate CpGs embedded in the primer region. The first HpaII site 5′ of the patch-methylated region is marked with an asterisk.
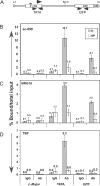
Figure 4. Promoter-Proximal Methylation Affects RNAPII Recruitment and TBP Binding
Chromatin isolated from unmethylated and patch-methylated clones 6− and 9P, respectively, was immunoprecipitated using antibodies (Abs) specific for the N-terminal domain of RNAPII (sc-899), the unphosphorylated CTD of RNAPII (8WG16), TBP, or control rabbit IgG. Quantitative real-time PCR was conducted using the transgene specific primers shown (A), or a control primer pair specific for the endogenous β-maj gene. The mean percent (+/− standard deviation) of bound material/total input chromatin for two to three independent chromatin preparations is shown for each ChIP experiment. No enrichment of RNAPII was detected in the silent β-maj gene in either clone with sc-899 (B) or 8WG16 (C), demonstrating the low level of background detected under the conditions used. Analysis of the TATA box and GFP regions of the transgene reveals significant enrichment of RNAPII exclusively in the unmethylated, highly transcribed clone, particularly at the 5′ end of the transgene. Similarly, TBP binding is detected exclusively in the promoter region of the unmethylated clone (D), suggesting that inhibition of transcription of the patch-methylated cassette occurs as the level of transcription initiation.
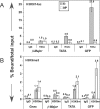
Figure 5. Promoter-Proximal DNA Methylation Leads to Hypoacetylation of H3K9/14 across the Transgene and Local H3K9 Trimethylation
Chromatin was generated from clones 6− and 9P, and ChIP was conducted using antibodies specific for H3 acetylated on K9/K14, H3 trimethylated on K9 or control rabbit IgG. Quantitative real-time PCR was carried out on the immunoprecipitated DNA using primers specific for the endogenous Gnas and/or β-maj genes as internal controls and the TATA box or GFP regions of the transgene (shown in Figure 4A). For each antibody, values shown represent the mean percent (+/− standard deviation) of bound material/total input chromatin for three independent experiments. (A) Analysis of the TATA box and GFP regions of the transgene reveals enrichment of H3K9/14Ac (H3Ac) exclusively in the highly transcribed unmethylated cassette. The level of enrichment of H3K9/14Ac in the β-maj gene promoter region was similar between the two clones, indicating that the immunoprecipitations worked with similar efficiencies. (B) Analysis of the TATA box and GFP regions of the transgene reveals a significantly higher level of H3K9me3 (H3K9m) enrichment exclusively in the GFP region of clone 9P. As this region shows a high density of DNA methylation, these results reveal that DNA methylation is sufficient to promote H3K9 trimethylation in this euchromatic region. The levels of enrichment of H3K9me3 at Gnas and β-maj was similar between the two clones, indicating that the immunoprecipitations worked with similar efficiencies.
Figure 6. Promoter-Proximal DNA Methylation Influences the Chromatin Structure of the Unmethylated Promoter Region
To compare the chromatin structure of the unmethylated versus patch-methylated transgenes, intact nuclei from clones 6− and 9P respectively, were digested with increasing concentrations of DNase I. Subsequently, genomic DNA was isolated and digested with BamHI. Digested DNA was resolved on a 0.8% agarose gel. A map of the transgene, GFP probe, and HS sites is shown (A). The location of transgene-specific DNase HS sites was determined via Southern hybridization using a GFP probe (B). Predicted HS sites at the proviral enhancer and promoter, 2.5 kb and 2.3 kb upstream of the BamHI site, respectively, are shown. Note that the promoter-specific HS site is absent in clone 9P, even in the presence of relatively high concentrations of DNase I. Two additional HS sites, one site that is common to both clones and maps to a genomic (g) site (within a Charlie4 DNA transposon) ∼3 kb upstream of the enhancer HS site, and a second that is unique to the 6− clone and maps to the splice acceptor (SA) site within the transgene (∼1.2 kb upstream of the BamHI site) were also detected. To demonstrate comparable efficiencies of DNase I digestion in the two clones, the blot shown in (B) was stripped and reprobed with a probe specific for the “HS3” hypersensitive site in the endogenous β-globin Locus Control Region (C).
Figure 7. Promoter-Proximal DNA Methylation Influences Nucleosome Positioning in the Unmethylated Promoter Region
To determine nucleosome positioning around the promoter region, nuclei from clones 6− and 9P were treated in parallel with M.SssI for 15 min. Subsequently, genomic DNA was isolated and analyzed by bisulphite sequencing using primers flanking the promoter/TSS region. For each clone, 24 molecules were cloned and sequenced. Methylated and unmethylated CpGs are depicted as filled and open ovals, respectively (A). The fraction of molecules harboring an unmethylated cytosine at each CpG in the sequenced region is plotted in (B). Note the dramatic difference in methylation efficiency spanning the region from ∼CpG #5 to ∼CpG #18 (a distance of 148 bps), particularly those sites flanking the TSS at ∼CpG #11. Based on these data, a diagram showing the predicted predominant sites of nucleosome occupancy (ovals) for each clone is shown. The enhancer region (CpGs #1–3) is likely to be constitutively nucleosome-free, consistent with the presence of a DNase HS site in this region in both clones.
Figure 8. Model of the Relationship between Promoter-Proximal DNA Methylation and Transcriptional Efficiency
Four alternative DNA methylation states are shown, along with the acetylation (K9ac) and methylation (K9me) states of H3K9. In the presence of “distal” DNA methylation (>1–2 kb from the TSS), transcription may not be effected. This would allow for methylation and silencing of intronic parasitic elements, such as SINE and LINES or “cryptic promoters”, while allowing for efficient transcription of bona fide genes. Dense DNA methylation on the order of 1 kb from the TSS may have a modest effect on elongation efficiency, perhaps by altering the chromatin structure of the promoter proximal region, as we have shown previously [19]. Promoter proximal methylation (∼300 bp from the TSS) may have a dramatic effect on transcription initiation, as a result of altering promoter chromatin structure, as described here. Methylation of the promoter region itself leads to robust silencing of the gene, as is frequently observed for CpG island promoters in cancer cells.
Similar articles
- DNA methylation directly silences genes with non-CpG island promoters and establishes a nucleosome occupied promoter.
Han H, Cortez CC, Yang X, Nichols PW, Jones PA, Liang G. Han H, et al. Hum Mol Genet. 2011 Nov 15;20(22):4299-310. doi: 10.1093/hmg/ddr356. Epub 2011 Aug 11. Hum Mol Genet. 2011. PMID: 21835883 Free PMC article. - Genomic targeting of methylated DNA: influence of methylation on transcription, replication, chromatin structure, and histone acetylation.
Schübeler D, Lorincz MC, Cimbora DM, Telling A, Feng YQ, Bouhassira EE, Groudine M. Schübeler D, et al. Mol Cell Biol. 2000 Dec;20(24):9103-12. doi: 10.1128/MCB.20.24.9103-9112.2000. Mol Cell Biol. 2000. PMID: 11094062 Free PMC article. - The expression of endothelial nitric-oxide synthase is controlled by a cell-specific histone code.
Fish JE, Matouk CC, Rachlis A, Lin S, Tai SC, D'Abreo C, Marsden PA. Fish JE, et al. J Biol Chem. 2005 Jul 1;280(26):24824-38. doi: 10.1074/jbc.M502115200. Epub 2005 May 3. J Biol Chem. 2005. PMID: 15870070 - Mammalian RNA polymerase II core promoters: insights from genome-wide studies.
Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Sandelin A, et al. Nat Rev Genet. 2007 Jun;8(6):424-36. doi: 10.1038/nrg2026. Epub 2007 May 8. Nat Rev Genet. 2007. PMID: 17486122 Review. - Divergent transcription: a new feature of active promoters.
Seila AC, Core LJ, Lis JT, Sharp PA. Seila AC, et al. Cell Cycle. 2009 Aug 15;8(16):2557-64. doi: 10.4161/cc.8.16.9305. Epub 2009 Aug 19. Cell Cycle. 2009. PMID: 19597342 Review.
Cited by
- Cis-regulatory sequences in plants: Their importance, discovery, and future challenges.
Schmitz RJ, Grotewold E, Stam M. Schmitz RJ, et al. Plant Cell. 2022 Feb 3;34(2):718-741. doi: 10.1093/plcell/koab281. Plant Cell. 2022. PMID: 34918159 Free PMC article. Review. - Mouse strain-specific polymorphic provirus functions as cis-regulatory element leading to epigenomic and transcriptomic variations.
Zhou X, Sam TW, Lee AY, Leung D. Zhou X, et al. Nat Commun. 2021 Nov 9;12(1):6462. doi: 10.1038/s41467-021-26630-z. Nat Commun. 2021. PMID: 34753915 Free PMC article. - An Analysis of Methylome Evolution in Primates.
Sahm A, Koch P, Horvath S, Hoffmann S. Sahm A, et al. Mol Biol Evol. 2021 Oct 27;38(11):4700-4714. doi: 10.1093/molbev/msab189. Mol Biol Evol. 2021. PMID: 34175932 Free PMC article. - SP8 Promotes an Aggressive Phenotype in Hepatoblastoma via FGF8 Activation.
Wagner AE, Schwarzmayr T, Häberle B, Vokuhl C, Schmid I, von Schweinitz D, Kappler R. Wagner AE, et al. Cancers (Basel). 2020 Aug 15;12(8):2294. doi: 10.3390/cancers12082294. Cancers (Basel). 2020. PMID: 32824198 Free PMC article. - Cytosine methylations in the promoter regions of genes involved in the cellular oxidation equilibrium pathways affect rice heat tolerance.
He C, Zhang HY, Zhang YX, Fu P, You LL, Xiao WB, Wang ZH, Song HY, Huang YJ, Liao JL. He C, et al. BMC Genomics. 2020 Aug 14;21(1):560. doi: 10.1186/s12864-020-06975-3. BMC Genomics. 2020. PMID: 32799794 Free PMC article.
References
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. - PubMed
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. - PubMed
- Goll MG, Bestor TH. Eukaryotic cytosine methyltransferases. Annu Rev Biochem. 2005;74:481–514. - PubMed
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases