The complete nucleotide sequence of the coffee (Coffea arabica L.) chloroplast genome: organization and implications for biotechnology and phylogenetic relationships amongst angiosperms - PubMed (original) (raw)
The complete nucleotide sequence of the coffee (Coffea arabica L.) chloroplast genome: organization and implications for biotechnology and phylogenetic relationships amongst angiosperms
Nalapalli Samson et al. Plant Biotechnol J. 2007 Mar.
Abstract
The chloroplast genome sequence of Coffea arabica L., the first sequenced member of the fourth largest family of angiosperms, Rubiaceae, is reported. The genome is 155 189 bp in length, including a pair of inverted repeats of 25,943 bp. Of the 130 genes present, 112 are distinct and 18 are duplicated in the inverted repeat. The coding region comprises 79 protein genes, 29 transfer RNA genes, four ribosomal RNA genes and 18 genes containing introns (three with three exons). Repeat analysis revealed five direct and three inverted repeats of 30 bp or longer with a sequence identity of 90% or more. Comparisons of the coffee chloroplast genome with sequenced genomes of the closely related family Solanaceae indicated that coffee has a portion of rps19 duplicated in the inverted repeat and an intact copy of infA. Furthermore, whole-genome comparisons identified large indels (> 500 bp) in several intergenic spacer regions and introns in the Solanaceae, including trnE (UUC)-trnT (GGU) spacer, ycf4-cemA spacer, trnI (GAU) intron and rrn5-trnR (ACG) spacer. Phylogenetic analyses based on the DNA sequences of 61 protein-coding genes for 35 taxa, performed using both maximum parsimony and maximum likelihood methods, strongly supported the monophyly of several major clades of angiosperms, including monocots, eudicots, rosids, asterids, eurosids II, and euasterids I and II. Coffea (Rubiaceae, Gentianales) is only the second order sampled from the euasterid I clade. The availability of the complete chloroplast genome of coffee provides regulatory and intergenic spacer sequences for utilization in chloroplast genetic engineering to improve this important crop.
Figures
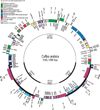
Figure 1
Circular gene map of the Coffea arabica chloroplast genome. The thick lines indicate the extent of the inverted repeats (IRa and IRb, 25 943 bp), which separate the genome into small (SSC, 18 133 bp) and large (LSC, 85 166 bp) single-copy regions. Genes on the outside of the map are transcribed in the clockwise direction and genes on the inside of the map are transcribed in the counterclockwise direction. *The rps19 gene locates entirely in the IRb region and partly in the IRa region. Arrows show the location of repeats (for more details on repeats, see Table 3).
Figure 2
The chloroplast genome comparison derived through a percentage identity plot of coffee against four Solanaceae members (Atropa belladonna, Solanum bulbocastanum, Nicotiana tabacum and Solanum lycopersicum) using the MultiPipMaker alignment tool. DNA losses are marked with roman numerals and the red boxes. I, region within intergenic spacer (IGS) [rps16_–_trnQ (UUG)]; II, region within IGS [trnE (UUC)–trnI (GGU)]; III, region within IGS (ycf4_–_cemA); IV, intron [IRb: trnI (GAU)]; V, intron [IRa: trnI (GAU)].
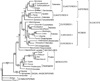
Figure 3
Maximum parsimony tree based on 61 chloroplast protein-coding genes (data are available at
http://www.biosci.utexas.edu/IB/faculty/jansen/lab/research/datafiles/index.htm
). The single most parsimonious phylogram has a length of 61 797, a consistency index of 0.41 (excluding uninformative characters) and a retention index of 0.58. Numbers above and below the nodes indicate the number of nucleotide substitutions and bootstrap support values, respectively.
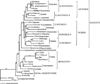
Figure 4
Maximum likelihood tree based on 61 chloroplast protein-coding genes. The single maximum likelihood phylogram has a maximum likelihood value of −ln L = 348 679.23765. Numbers at the nodes indicate the bootstrap support values and the branch length scale is shown at the base of the tree.
Similar articles
- The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var 'Ridge Pineapple': organization and phylogenetic relationships to other angiosperms.
Bausher MG, Singh ND, Lee SB, Jansen RK, Daniell H. Bausher MG, et al. BMC Plant Biol. 2006 Sep 30;6:21. doi: 10.1186/1471-2229-6-21. BMC Plant Biol. 2006. PMID: 17010212 Free PMC article. - Complete plastid genome sequence of Daucus carota: implications for biotechnology and phylogeny of angiosperms.
Ruhlman T, Lee SB, Jansen RK, Hostetler JB, Tallon LJ, Town CD, Daniell H. Ruhlman T, et al. BMC Genomics. 2006 Aug 31;7:222. doi: 10.1186/1471-2164-7-222. BMC Genomics. 2006. PMID: 16945140 Free PMC article. - The complete chloroplast genome sequence of Gossypium hirsutum: organization and phylogenetic relationships to other angiosperms.
Lee SB, Kaittanis C, Jansen RK, Hostetler JB, Tallon LJ, Town CD, Daniell H. Lee SB, et al. BMC Genomics. 2006 Mar 23;7:61. doi: 10.1186/1471-2164-7-61. BMC Genomics. 2006. PMID: 16553962 Free PMC article. - Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids.
Jansen RK, Kaittanis C, Saski C, Lee SB, Tomkins J, Alverson AJ, Daniell H. Jansen RK, et al. BMC Evol Biol. 2006 Apr 9;6:32. doi: 10.1186/1471-2148-6-32. BMC Evol Biol. 2006. PMID: 16603088 Free PMC article. - Comparative genomics of four Liliales families inferred from the complete chloroplast genome sequence of Veratrum patulum O. Loes. (Melanthiaceae).
Do HD, Kim JS, Kim JH. Do HD, et al. Gene. 2013 Nov 10;530(2):229-35. doi: 10.1016/j.gene.2013.07.100. Epub 2013 Aug 23. Gene. 2013. PMID: 23973725
Cited by
- Characterisation of the complete chloroplast genome of Solanum tuberosum cv. White Lady.
Frank K, Nagy E, Taller J, Wolf I, Polgár Z. Frank K, et al. Biol Futur. 2024 Dec;75(4):401-410. doi: 10.1007/s42977-024-00240-4. Epub 2024 Sep 9. Biol Futur. 2024. PMID: 39251554 - The complete chloroplast genome of Gardenia stenophylla Merr (Rubiaceae) and its phylogenetic analysis.
Deng S, Fan C, Lu Z, Yang H. Deng S, et al. Mitochondrial DNA B Resour. 2024 Aug 12;9(8):1039-1043. doi: 10.1080/23802359.2024.2389918. eCollection 2024. Mitochondrial DNA B Resour. 2024. PMID: 39139657 Free PMC article. - Plastid phylogenomics and cytonuclear discordance in Rubioideae, Rubiaceae.
Thureborn O, Wikström N, Razafimandimbison SG, Rydin C. Thureborn O, et al. PLoS One. 2024 May 20;19(5):e0302365. doi: 10.1371/journal.pone.0302365. eCollection 2024. PLoS One. 2024. PMID: 38768140 Free PMC article. - Phylogenomics and topological conflicts in the tribe Anthospermeae (Rubiaceae).
Thureborn O, Wikström N, Razafimandimbison SG, Rydin C. Thureborn O, et al. Ecol Evol. 2024 Jan 25;14(1):e10868. doi: 10.1002/ece3.10868. eCollection 2024 Jan. Ecol Evol. 2024. PMID: 38274863 Free PMC article. - The chloroplast protein HCF164 is predicted to be associated with Coffea SH9 resistance factor against Hemileia vastatrix.
Guerra-Guimarães L, Pinheiro C, Oliveira ASF, Mira-Jover A, Valverde J, Guedes FAF, Azevedo H, Várzea V, Muñoz Pajares AJ. Guerra-Guimarães L, et al. Sci Rep. 2023 Sep 25;13(1):16019. doi: 10.1038/s41598-023-41950-4. Sci Rep. 2023. PMID: 37749157 Free PMC article.
References
- Asano T, Tsudzuki T, Takahashi S, Shimada H, Kadowaki K. Nucleotide sequence of the sugarcane (Saccharum officinarum) chloroplast genome: a comparative analysis of four monocot chloroplast genomes. DNA Res. 2004;11:93–99. - PubMed
- Ashihara H, Crozier A. Caffeine: a well known but little mentioned compound in plant science. Trends Plant Sci. 2001;6:407–413. - PubMed
- Barton C, Adam TL, Zaarowitz MA. Stable transformation of foreign DNA into Coffea arabica plants. 14th International Conference on Coffee Science; San Francisco, CA, USA. Paris: ASIC (Association Scientifique Internationale du Café); 1991. pp. 460–464.
- Carneiro M. Coffee biotechnology and its application in genetic transformation. Euphytica. 1997;96:167–172.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous