The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth - PubMed (original) (raw)
The Trithorax group protein Lid is a trimethyl histone H3K4 demethylase required for dMyc-induced cell growth
Julie Secombe et al. Genes Dev. 2007.
Abstract
The Myc oncoprotein is a potent inducer of cell growth, cell cycle progression, and apoptosis. While many direct Myc target genes have been identified, the molecular determinants of Myc's transcriptional specificity remain elusive. We have carried out a genetic screen in Drosophila and identified the Trithorax group protein Little imaginal discs (Lid) as a regulator of dMyc-induced cell growth. Lid binds to dMyc and is required for dMyc-induced expression of the growth regulatory gene Nop60B. The mammalian Lid orthologs, Rbp-2 (JARID1A) and Plu-1 (JARID1B), also bind to c-Myc, indicating that Lid-Myc function is conserved. We demonstrate that Lid is a JmjC-dependent trimethyl H3K4 demethylase in vivo and that this enzymatic activity is negatively regulated by dMyc, which binds to Lid's JmjC domain. Because Myc binding is associated with high levels of trimethylated H3K4, we propose that the Lid-dMyc complex facilitates Myc binding to, or maintenance of, this chromatin context.
Figures
Figure 1.
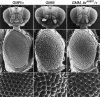
The GMM phenotype is suppressed by reducing the gene dose of lid. Scanning electron micrographs of adult eyes. (A,D,G) Control GMR-Gal4 flies. (B,E,H) GMM flies. (C,F,I) GMM, lidK6801/+ flies. A_–_C are 70× magnification, D_–_F show lateral views (170×), and G_–_I are 1000× magnification.
Figure 2.
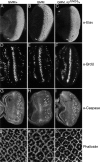
The GMM phenotype is primarily due to increased cell growth. Control GMR-Gal4/+ (A,D,G,J), GMM (B,E,H,K), and GMM, lid10424/+ (C,F,I,L) eye imaginal discs from wandering third instar larvae. (A_–_C) Anti-Elav to detect differentiating neurons posterior to the morphogenetic furrow (MF; marked with white bar). (D_–_F) BrdU incorporation to visualize cells in S phase. (G_–_I) Cell death visualized by expression of cleaved Caspase-3. (J_–_L) Cortical actin visualized by phalloidin-Rhodamine conjugate. Eye discs are orientated with anterior to the left.
Figure 3.
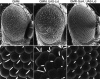
Increasing Lid levels enhances the GMM phenotype. Scanning electron micrographs of GMM (A,D), GMM, UAS-Lid (B,E), and GMR-Gal4, UAS-Lid (C,F) adult eyes. A_–_C show lateral views (170× magnification) and D_–_F show1000× magnification.
Figure 4.
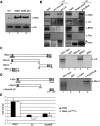
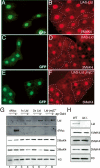
Lid physically interacts with dMyc. (A) Western blot of wild-type (lane 1), GMM (lane 2), and GMM heterozygous for lid10424 (lane 3) using 24 eye discs per lane. Top panel was probed with anti-dMyc; bottom panel shows anti-Histone H3 loading control. (B) Anti-Lid (left panel) and anti-dMyc (right panel) immunoprecipitations and Western blots for dMyc, Lid, Brm, Osa, and Ash2. A high-molecular-weight Ash2 is detected in both anti-Lid and anti-dMyc immunoprecipitates (arrow). (*) A nonspecific band. (C) Lid interacts with the C-terminal region of dMyc in vitro. In vitro assay testing binding of 35S-methionine Lid to GST (lane 2), GST-dMycN (lane 3), GST-dMycC (lane 4), or GST-dMax (lane 5). (D) In vitro binding assay using 35S-methionine-labeled Rbp-2 and GST (lane 2), c-mycN (lane 3), and c-mycbHLHZ (lane 4). (E) Real-time PCR analysis of dmyc, lid, and Nop60B expression levels relative to expression in GMR/+ controls in eye discs from GMM or GMM discs heterozygous for lid10424. Asterisk indicates that Nop60B levels in GMM and GMM heterozygous for lid are significantly different (Student’s _t_-test; P = 0.003).
Figure 5.
Lid is a trimethyl H3K4 demethylase. (A_–_F) Flp/Gal4 was used to clonally express Lid (A–D) or Lid-JmjC* (E,F) in fat body cells to examine di- and trimethylated H3K4 levels (red). (A,C,E) Clones expressing Lid or Lid-JmjC* (full-length Lid protein containing two point mutations in the demethylase domain) are marked by coexpression of GFP, and these clones are outlined in the other panels. (G) Western blots from third instar larval wing discs from control (−) or apterous-Gal4 (+) and UAS-dMyc (lanes 1,2), one copy of UAS-Lid (lanes 3,4), two copies of UAS-Lid (lanes 5,6), or UAS-Lid-JmjC* (lanes 7,8). Westerns were probed with anti-Lid, anti-dMyc, anti-dimethyl H3K4, anti-trimethyl H3K4, or a total histone H3 control. Two wing discs were used per lane for all Westerns except 3MeK4, in which four were used. (H) Western analysis of wild-type (left) and lid10424 homozygous mutant (right) wing discs using anti-Lid, anti-monomethyl H3K4, anti-dimethyl H3K4, anti-trimethyl H3K4, or total histone H3.
Figure 6.
Expression of Lid-JmjC* (full-length Lid protein containing two point mutations in the demethylase domain) enhances the GMM phenotype. (A–E) Scanning electron micrographs of GMM (A), GMM carrying two copies of UAS-Lid (B), GMM, UAS-Lid-JmjC* (C), and GMM; ago4/+ (D). All scanning electron micrographs are 170× magnification.
Figure 7.
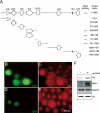
dMyc inhibits the demethylase activity of Lid. (A) Summary of GST fusion protein-mediated in vitro binding assays to map the region of Lid bound by dMyc. (+++) Strong binding (∼20% input); (−) no detectable binding. (B–E) Flp/Gal4-mediated clones coexpressing Lid and dMyc marked by GFP expression (outlined in C,E) and stained for Lid (C) or trimethyl H3K4 (E). (H) Western analysis from UAS-Lid, UAS-dMyc (−), or apterous-Gal4, UAS-Lid, UAS-dMyc (+) wing discs. Westerns were probed with anti-Lid, anti-dMyc, anti-trimethyl H3K4, and total histone H3. Four wing discs were used per lane.
Similar articles
- Essential functions of the histone demethylase lid.
Li L, Greer C, Eisenman RN, Secombe J. Li L, et al. PLoS Genet. 2010 Nov 24;6(11):e1001221. doi: 10.1371/journal.pgen.1001221. PLoS Genet. 2010. PMID: 21124823 Free PMC article. - Coordinated regulation of Myc trans-activation targets by Polycomb and the Trithorax group protein Ash1.
Goodliffe JM, Cole MD, Wieschaus E. Goodliffe JM, et al. BMC Mol Biol. 2007 May 22;8:40. doi: 10.1186/1471-2199-8-40. BMC Mol Biol. 2007. PMID: 17519021 Free PMC article. - The trithorax-group gene in Drosophila little imaginal discs encodes a trimethylated histone H3 Lys4 demethylase.
Eissenberg JC, Lee MG, Schneider J, Ilvarsonn A, Shiekhattar R, Shilatifard A. Eissenberg JC, et al. Nat Struct Mol Biol. 2007 Apr;14(4):344-6. doi: 10.1038/nsmb1217. Epub 2007 Mar 11. Nat Struct Mol Biol. 2007. PMID: 17351630 - Myc--what we have learned from flies.
Siddall NA, Lin JI, Hime GR, Quinn LM. Siddall NA, et al. Curr Drug Targets. 2009 Jul;10(7):590-601. doi: 10.2174/138945009788680400. Curr Drug Targets. 2009. PMID: 19601763 Review. - The function and regulation of the JARID1 family of histone H3 lysine 4 demethylases: the Myc connection.
Secombe J, Eisenman RN. Secombe J, et al. Cell Cycle. 2007 Jun 1;6(11):1324-8. doi: 10.4161/cc.6.11.4269. Epub 2007 Jun 14. Cell Cycle. 2007. PMID: 17568193 Review.
Cited by
- Identification of small molecule inhibitors of Jumonji AT-rich interactive domain 1B (JARID1B) histone demethylase by a sensitive high throughput screen.
Sayegh J, Cao J, Zou MR, Morales A, Blair LP, Norcia M, Hoyer D, Tackett AJ, Merkel JS, Yan Q. Sayegh J, et al. J Biol Chem. 2013 Mar 29;288(13):9408-17. doi: 10.1074/jbc.M112.419861. Epub 2013 Feb 13. J Biol Chem. 2013. PMID: 23408432 Free PMC article. - Histone demethylase RBP2 decreases miR-21 in blast crisis of chronic myeloid leukemia.
Zhou M, Zeng J, Wang X, Wang X, Huang T, Fu Y, Sun T, Jia J, Chen C. Zhou M, et al. Oncotarget. 2015 Jan 20;6(2):1249-61. doi: 10.18632/oncotarget.2859. Oncotarget. 2015. PMID: 25575817 Free PMC article. - Selective targeting of histone methylation.
Islam AB, Richter WF, Lopez-Bigas N, Benevolenskaya EV. Islam AB, et al. Cell Cycle. 2011 Feb 1;10(3):413-24. doi: 10.4161/cc.10.3.14705. Epub 2011 Feb 1. Cell Cycle. 2011. PMID: 21270517 Free PMC article. - dSet1 is the main H3K4 di- and tri-methyltransferase throughout Drosophila development.
Hallson G, Hollebakken RE, Li T, Syrzycka M, Kim I, Cotsworth S, Fitzpatrick KA, Sinclair DA, Honda BM. Hallson G, et al. Genetics. 2012 Jan;190(1):91-100. doi: 10.1534/genetics.111.135863. Epub 2011 Nov 2. Genetics. 2012. PMID: 22048023 Free PMC article. - Epigenetic regulator Lid maintains germline stem cells through regulating JAK-STAT signaling pathway activity.
Tarayrah L, Li Y, Gan Q, Chen X. Tarayrah L, et al. Biol Open. 2015 Oct 21;4(11):1518-27. doi: 10.1242/bio.013961. Biol Open. 2015. PMID: 26490676 Free PMC article.
References
- Aasland R., Gibson T.J., Stewart A.F., Gibson T.J., Stewart A.F., Stewart A.F. The PHD finger—Implications for chromatin-mediated transcriptional regulation. Trends Biochem. Sci. 1995;20:56–59. - PubMed
- Adhikary S., Eilers M., Eilers M. Transcriptional regulation and transformation by MYC proteins. Nat. Rev. Mol. Cell Biol. 2005;6:635–645. - PubMed
- Amati B., Frank S.R., Donjerkovic D., Taubert S., Frank S.R., Donjerkovic D., Taubert S., Donjerkovic D., Taubert S., Taubert S. Function of the c-Myc oncoprotein in chromatin remodeling and transcription. Biochim. Biophys. Acta. 2001;1471:M135–M145. - PubMed
- Angulo M., Corominas M., Serras F., Corominas M., Serras F., Serras F. Activation and repression activities of ash2 in Drosophila wing imaginal discs. Development. 2004;131:4943–4953. - PubMed
- Arabi A., Wu S.Q., Ridderstrale K., Bierhoff H., Shiue C., Fatyol K., Fahlen S., Hydbring P., Soderberg O., Grummt I., Wu S.Q., Ridderstrale K., Bierhoff H., Shiue C., Fatyol K., Fahlen S., Hydbring P., Soderberg O., Grummt I., Ridderstrale K., Bierhoff H., Shiue C., Fatyol K., Fahlen S., Hydbring P., Soderberg O., Grummt I., Bierhoff H., Shiue C., Fatyol K., Fahlen S., Hydbring P., Soderberg O., Grummt I., Shiue C., Fatyol K., Fahlen S., Hydbring P., Soderberg O., Grummt I., Fatyol K., Fahlen S., Hydbring P., Soderberg O., Grummt I., Fahlen S., Hydbring P., Soderberg O., Grummt I., Hydbring P., Soderberg O., Grummt I., Soderberg O., Grummt I., Grummt I., et al. c-Myc associates with ribosomal DNA and activates RNA polymerase I transcription. Nat. Cell Biol. 2005;7:303–310. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous