Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella - PubMed (original) (raw)
Functional analysis of an individual IFT protein: IFT46 is required for transport of outer dynein arms into flagella
Yuqing Hou et al. J Cell Biol. 2007.
Abstract
Intraflagellar transport (IFT), which is the bidirectional movement of particles within flagella, is required for flagellar assembly. IFT particles are composed of approximately 16 proteins, which are organized into complexes A and B. We have cloned Chlamydomonas reinhardtii and mouse IFT46, and show that IFT46 is a highly conserved complex B protein in both organisms. A C. reinhardtii insertional mutant null for IFT46 has short, paralyzed flagella lacking dynein arms and with central pair defects. The mutant has greatly reduced levels of most complex B proteins, indicating that IFT46 is necessary for complex B stability. A partial suppressor mutation restores flagellar length to the ift46 mutant. IFT46 is still absent, but levels of the other IFT particle proteins are largely restored, indicating that complex B is stabilized in the suppressed strain. Axonemal ultrastructure is restored, except that the outer arms are still missing, although outer arm subunits are present in the cytoplasm. Thus, IFT46 is specifically required for transporting outer arms into the flagellum.
Figures
Figure 1.
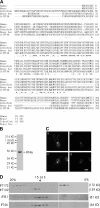
Identification and characterization of C. reinhardtii IFT46. (A) Predicted amino acid sequence of IFT46 and alignment with orthologues from other organisms. CrIFT46 is highly conserved from aa 100–315. Amino acid identities are marked with asterisks; similarities are marked with either one or two dots. Sequences used in this alignment are as follows: human (AAH22856), mouse (NP_076320), zebrafish (XP_694278), honey bee (XP_396519), and CrIFT46 (DQ787426). (B) The antibody to IFT46 specifically recognizes a ∼46-kD doublet in Western blots of whole cell lysates; the doublet is caused by phosphorylation (unpublished data). (C) IFT46 has a typical cellular localization for an IFT particle protein. Cells were labeled with the anti-IFT46 antibody. Images of the same cell were acquired focusing at the flagella (a and c), or at the basal body region (b and d) with much less exposure time. The majority of IFT46 is located at the peribasal body region, with a lesser amount distributed along the flagella as distinct dots. Bar, 5 μm. (D) IFT46 comigrates with IFT81, an IFT complex B protein, in sucrose gradients. The flagellar membrane plus matrix was fractionated in a 5–20% sucrose gradient. The fractions were analyzed by Western blotting using antibodies to IFT46, complex B proteins IFT172 and IFT81, and complex A protein IFT139. Under these experimental conditions, complex B separated from complex A, and IFT172 dissociated from complex B.
Figure 2.
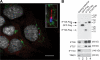
The mouse orthologue of CrIFT46 is also an IFT complex B protein. (A) MmIFT46 localizes in cilia. IMCD3 cells expressing Flag-tagged MmIFT46 were labeled with antibodies to Flag (red), IFT20 (green), which localizes to Golgi in addition to cilia, and a centrosome component (blue). Nuclei were DAPI stained and are shown as gray. IFT46-Flag was concentrated at the bases of the cilia, but was also located along the length of the cilia. The inset shows an enlargement of one of the cilia. Bar, 10 μm. (B) IFT complex B proteins coimmunoprecipitate with MmIFT46. Whole-cell lysates from mouse IMCD3 cells expressing Flag-tagged GFP (lane 2), Flag-tagged IFT20 (lane 3), or Flag-tagged IFT46 (lane 4) were immunoprecipitated with an anti-Flag antibody. The immunoprecipates were analyzed by Western blotting with antibodies to Flag (top), IFT88, IFT57, IFT140, and TIM23, which is a mitochondrial protein used as a control (bottom). (top) The positions of the Flag-tagged proteins are indicated with arrows. *, an irrelevant protein that is recognized by the Flag antibody. Lane 1 is the starting material for the IFT46-Flag immunoprecipitation. Note that IFT46-Flag is highly enriched by the immunoprecipitation (compare lanes 1 and 4). Lanes were loaded with equivalent amounts of protein, except that in the top gel only, lane 2 was loaded with 1/50 the amount of protein that is present in the other lanes. Complex B proteins IFT88 and IFT57 coimmunoprecipitated with MmIFT46-FLAG (and MmIFT20-FLAG), whereas complex A protein IFT140 and TIM23 did not, even though they were present in the starting material. No IFT proteins were coimmunoprecipated from the lysate of the Flag-GFP–expressing cells.
Figure 3.
The nonmotile phenotype of YH6 is caused by a null mutation in the IFT46 gene. (A) IFT46 gene structure. There are two PstI sites (P) within the IFT46 gene. Three primer sets used for PCR to map the mutation region in the mutant are indicated by arrowheads. Regions corresponding to the ∼4.8-kb genomic fragment used for rescue experiments, the cDNA probe used for screening the mutant collection, and the genomic DNA probe used for Southern blotting also are shown. (B) PCR results using primers to different regions of the IFT46 gene showed that strain T8a4-11 is defective in the middle portion of the gene. (C and D) The nonmotile phenotype of YH6, which is an offspring of a cross between T8a4-11 and wild-type cells, was rescued by transforming the mutant cells with the wild-type IFT46 gene. Genomic DNAs from wild-type cells, YH6, and rescued cells were cut by PstI and analyzed by Southern blotting using a IFT46 genomic DNA probe (C). The rescued cells (6R29 through 6R38) have the 1.5-kb fragment originating from the transgene (the 1.9-kb fragment is missing because it was not present in its entirety in the transgene) and two hybridizing bands (*) originating from the mutated ift46 gene. They also have other hybridizing bands, indicating that the IFT46 transgenes were incorporated at different sites. Western blots of whole cell lysates (D) show that YH6 cells lack the IFT46 protein and that the rescued cells express IFT46 protein. The same blot was stripped and probed with an anti-tubulin antibody as control (bottom).
Figure 4.
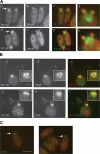
Flagellar assembly is defective in ift46 mutant cells. (A) Electron micrographs show that ift46 flagella are short with normal basal bodies and transition zones. The mutant flagella lack dynein arms, and central pair assembly is defective. (a–c) Cross sections of wild-type (WT) flagella. (d) Cross section of a flagellum of the rescued cell line 6R32 (R). (e) Longitudinal section of a flagellum of 6R32. (f–k) Cross sections of ift46 flagella (M). (l–p) Longitudinal sections of ift46 flagella (M). The membrane vesicles or bulges frequently present at the tips of the short mutant flagella are also observed in the ift88 mutant (Pazour et al., 2000). Bars, 100 nm. (B) Western blotting shows that ift46 flagella lack outer dynein arm and inner dynein arm components, although these components exist in the cell cytoplasm. In contrast, the outer dynein arm docking complex is transported into ift46 flagella. Flagellar samples (Fla) and whole cell lysates (Cell) from ift46 or wild-type cells were probed with antibodies to IC2 (an outer dynein arm component), IC138 (an inner dynein arm I1 component), or DC2 (a subunit of the outer dynein arm docking complex). Tubulin was probed as a loading control.
Figure 5.
IFT complex B is unstable in the ift46 mutant, but stabilized in the partially suppressed strain. Whole-cell lysates from wild-type cells, Supift461 cells, and ift46 cells were analyzed in Western blots probed with antibodies to several IFT particle proteins, IC2, and DC2. Complex A protein levels are increased in ift46 compared with wild-type cells. With the exception of IFT172, complex B protein levels are dramatically decreased in ift46 compared with wild-type cells. The levels of these proteins in Supift461 cells are between those in ift46 and wild-type cells. IFT172 is increased in Supift461 cells. Tubulin was used to normalize the loading of the samples. Note that both ift46 cells and Supift461 cells have IC2 and DC2.
Figure 6.
Complex A and B proteins differ in cellular distribution, and loss of IFT46 affects the cell body localization of IFT172, but not of IFT139. (A) ift46 cells were double labeled either with antibodies to IFT172 (a) and tubulin (b) or with antibodies to IFT139 (e) and tubulin (f). c and g are merged images; d and h are enlargements of the flagella and basal body regions. IFT172 and IFT139 are both present in the short flagella (arrows). In the basal body region, IFT139 localizes more anteriorly than IFT172. (B) Wild-type cells were double-labeled either with antibodies to IFT172 (a) and IFT46 (b) or with antibodies to IFT139 (d) and IFT46 (e). The merged images are shown in c and f; the insets show enlargements of the basal body regions. IFT172 usually colocalizes with IFT46. However, IFT139 usually only partially colocalizes with IFT46 in the anterior part of the basal body region. (C) In the absence of IFT46, the remaining complex B proteins are still transported into the flagella. ift46 cells were labeled with an antibody to IFT57, which is located in flagella (arrows), as well as in the basal body region. Bars, 5 μm.
Figure 7.
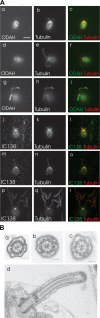
The partial suppressor of ift46 has defects in assembly of the outer dynein arm, but not inner dynein arm I1. (A) Cells were double labeled either with antibodies to outer dynein arm heavy chain α (a, d, and g) and tubulin (b, e, and h), or with antibodies to inner dynein arm intermediate chain IC138 (j, m, and p) and tubulin (k, n, and q). The right column shows the merged images. a–f and j–o are Supift461 cells; g–i and p–r are wild-type cells. Note that Supift461 cells have flagella of varying lengths. (B) Electron micrographs of wild-type flagella (a) and Supift461 flagella (b–d). Supift461 flagella lack outer arms, but the axonemal ultrastructure is otherwise normal. Bars: (A) 5 μm; (B) 100 nm.
Figure 8.
The 3′ end of the IFT46 gene is expressed in the suppressor, but not in the ift46 mutant. RT-PCR was performed with no added DNA (control, C), or cDNA isolated from wild-type cells (WT), ift46 cells without aeration (_ift46_s), ift46 cells with aeration (ift46), Supift461 cells without aeration (Sups), and Supift461 cells with aeration (Sup) using primers designed to amplify transcripts from the 5′, middle, and 3′ regions of the IFT46 gene. The products were then electrophoresed in 1.5% agarose gels. The results show that the 5′ portion of the IFT46 gene is transcribed in all the cells. The middle region is not transcribed in either the ift46 mutant cells or the suppressed cells. The 3′ region is not transcribed in the ift46 mutant cells, but is transcribed in the suppressed cells. The transcription of the 3′ region of the IFT46 gene is not dependent on the growth condition. This result implies that the suppression is caused by expression of the 3′ region of the IFT46 gene.
Similar articles
- The N-terminus of IFT46 mediates intraflagellar transport of outer arm dynein and its cargo-adaptor ODA16.
Hou Y, Witman GB. Hou Y, et al. Mol Biol Cell. 2017 Sep 1;28(18):2420-2433. doi: 10.1091/mbc.E17-03-0172. Epub 2017 Jul 12. Mol Biol Cell. 2017. PMID: 28701346 Free PMC article. - ODA16 aids axonemal outer row dynein assembly through an interaction with the intraflagellar transport machinery.
Ahmed NT, Gao C, Lucker BF, Cole DG, Mitchell DR. Ahmed NT, et al. J Cell Biol. 2008 Oct 20;183(2):313-22. doi: 10.1083/jcb.200802025. Epub 2008 Oct 13. J Cell Biol. 2008. PMID: 18852297 Free PMC article. - Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body.
Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL. Qin H, et al. J Cell Biol. 2004 Jan 19;164(2):255-66. doi: 10.1083/jcb.200308132. Epub 2004 Jan 12. J Cell Biol. 2004. PMID: 14718520 Free PMC article. - The intraflagellar transport machinery of Chlamydomonas reinhardtii.
Cole DG. Cole DG. Traffic. 2003 Jul;4(7):435-42. doi: 10.1034/j.1600-0854.2003.t01-1-00103.x. Traffic. 2003. PMID: 12795688 Review. - Regulation of flagellar length in Chlamydomonas.
Wilson NF, Iyer JK, Buchheim JA, Meek W. Wilson NF, et al. Semin Cell Dev Biol. 2008 Dec;19(6):494-501. doi: 10.1016/j.semcdb.2008.07.005. Epub 2008 Jul 25. Semin Cell Dev Biol. 2008. PMID: 18692148 Free PMC article. Review.
Cited by
- The Intraflagellar Transport Machinery.
Taschner M, Lorentzen E. Taschner M, et al. Cold Spring Harb Perspect Biol. 2016 Oct 3;8(10):a028092. doi: 10.1101/cshperspect.a028092. Cold Spring Harb Perspect Biol. 2016. PMID: 27352625 Free PMC article. Review. - Subunit interactions and organization of the Chlamydomonas reinhardtii intraflagellar transport complex A proteins.
Behal RH, Miller MS, Qin H, Lucker BF, Jones A, Cole DG. Behal RH, et al. J Biol Chem. 2012 Apr 6;287(15):11689-703. doi: 10.1074/jbc.M111.287102. Epub 2011 Dec 14. J Biol Chem. 2012. PMID: 22170070 Free PMC article. - Genetic and phenotypic analysis of flagellar assembly mutants in Chlamydomonas reinhardtii.
Iomini C, Till JE, Dutcher SK. Iomini C, et al. Methods Cell Biol. 2009;93:121-43. doi: 10.1016/S0091-679X(08)93007-7. Epub 2009 Dec 4. Methods Cell Biol. 2009. PMID: 20409815 Free PMC article. - Function and dynamics of PKD2 in Chlamydomonas reinhardtii flagella.
Huang K, Diener DR, Mitchell A, Pazour GJ, Witman GB, Rosenbaum JL. Huang K, et al. J Cell Biol. 2007 Nov 5;179(3):501-14. doi: 10.1083/jcb.200704069. J Cell Biol. 2007. PMID: 17984324 Free PMC article. - Intraflagellar transport particle size scales inversely with flagellar length: revisiting the balance-point length control model.
Engel BD, Ludington WB, Marshall WF. Engel BD, et al. J Cell Biol. 2009 Oct 5;187(1):81-9. doi: 10.1083/jcb.200812084. J Cell Biol. 2009. PMID: 19805630 Free PMC article.
References
- Ahmed, N.T., B. Lucker, D.G. Cole, and D.R. Mitchell. 2006. The Chlamydomonas ODA16 locus, needed for outer arm dynein assembly, encodes and intraflagellar transport adaptor. 46th Annual Meeting of the American Society for Cell Biology. Abstr. 1612.
- Brazelton, W.J., C.D. Amundsen, C.D. Silflow, and P.A. Lefebvre. 2001. The bld1 mutation identifies the Chlamydomonas osm-6 homolog as a gene required for flagellar assembly. Curr. Biol. 11:1591–1594. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R37 GM030626/GM/NIGMS NIH HHS/United States
- P30 DK032520/DK/NIDDK NIH HHS/United States
- R01 GM014642/GM/NIGMS NIH HHS/United States
- G30626/PHS HHS/United States
- P30 DK32520/DK/NIDDK NIH HHS/United States
- R37 GM014642/GM/NIGMS NIH HHS/United States
- GM60992/GM/NIGMS NIH HHS/United States
- R01 GM060992/GM/NIGMS NIH HHS/United States
- GM14642/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous