DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells - PubMed (original) (raw)
DNMT1 but not its interaction with the replication machinery is required for maintenance of DNA methylation in human cells
Fabio Spada et al. J Cell Biol. 2007.
Abstract
DNA methylation plays a central role in the epigenetic regulation of gene expression in vertebrates. Genetic and biochemical data indicated that DNA methyltransferase 1 (Dnmt1) is indispensable for the maintenance of DNA methylation patterns in mice, but targeting of the DNMT1 locus in human HCT116 tumor cells had only minor effects on genomic methylation and cell viability. In this study, we identified an alternative splicing in these cells that bypasses the disrupting selective marker and results in a catalytically active DNMT1 protein lacking the proliferating cell nuclear antigen-binding domain required for association with the replication machinery. Using a mechanism-based trapping assay, we show that this truncated DNMT1 protein displays only twofold reduced postreplicative DNA methylation maintenance activity in vivo. RNA interference-mediated knockdown of this truncated DNMT1 results in global genomic hypomethylation and cell death. These results indicate that DNMT1 is essential in mouse and human cells, but direct coupling of the replication of genetic and epigenetic information is not strictly required.
Figures
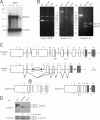
Figure 1.
_DNMT1_−/− (MT1KO) and DNMT1_−/−; DNMT3B_−/− (DKO) HCT116 cell lines express an internally deleted DNMT1 variant. (A) Northern blot analysis of HCT116 cell lines, including parental cells (+/+), two independent DNMT1 +/− clones, and one DNMT1 −/− clone (MT1KO). The blot was hybridized with a DNMT1 cDNA probe and a glyceraldehyde-3-phosphate dehydrogenase (GAPDH) probe as a loading control. The positions of the two transcripts and molecular weight markers are indicated. (B) RT-PCR analysis of parental (WT), DNMT1 −/− (MT1KO), and DNMT1 −/− ; DNMT3B −/− (DKO) HCT116 cells. The amplified regions are indicated at the bottom of each panel. In the left panel, the arrowhead indicates the specific PCR fragment, and the asterisk indicates an unspecific product. The right panel shows a nested PCR, where PCR products shown in the middle panel were used as templates. The two alternative splice forms expressed in MT1KO and DKO cells (right) and the positions of molecular weight markers are indicated. (C) Schematic drawing of the wt (top) and targeted DNMT1 alleles (middle) and alternatively spliced transcripts from the latter allele (bottom). Exons are shown as open rectangles (numbered on top) and are drawn to scale, whereas noncoding sequences are shown as lines and are not in scale. Sequences coding for major peptide domains are shown in different shades of gray, and their names are reported at the bottom. The positions of primers used for RT-PCR, translational start (ATG) codons, and termination (STOP) codons are indicated. (D) Western blot analysis of parental HCT116 cells and DNMT KO derivatives with antibodies to DNMT1 and lamin B1 (as a loading control). The molecular mass of major bands is indicated. The 170-kD band detected with the anti-DNMT1 antibody in the parental cells lane has a higher molecular mass than the band detected in MT1KO and DKO lanes and likely represents a DNMT1 degradation product. The percentages of normalized signal levels from the DNMT1 variant in MT1KO and DKO cells with respect to wt DNMT1 levels in HCT116 cells (100%) are shown at the bottom of the corresponding lanes.
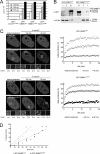
Figure 2.
GFP-DNMT1ΔE3–6 is catalytically active in vitro and is capable of postreplicative DNA methylation in vivo. (A) In vitro DNMT assay of GFP-DNMT1 fusion variants transiently expressed in HEK293T cells and immunopurified with an anti-GFP antibody. (B) Coimmunoprecipitation of endogenous PCNA with either GFP-DNMT1wt or GFP-DNMT1ΔE3–6 expressed in HEK293T cells. Input (I), flow through (F), and bound (B) fractions are shown; 5% of input and flow through fractions with respect to bound fractions was loaded. The molecular masses of the two GFP fusions are indicated on the right. (C and D) Trapping assay for GFP-DNMT1wt and GFP-DNMT1ΔE3–6 transiently expressed in HeLa cells. (C) Images on the left show cells in early S phase according to the RFP-PCNA pattern and expressing the indicated GFP-DNMT1 variant. The distribution of fluorescent fusions is shown before and at indicated time points after the addition of 30 μM 5-aza-dC. Regions targeted by photobleaching are indicated by boxes and are shown magnified at the bottom before and 0.3 (first postbleaching time point) and 20 s after bleaching. Bars, 5 μm. Plots on the right show FRAP curves of GFP fusions at targeted regions before and at indicated time points after the addition of 5-aza-dC from the corresponding cells shown on the left. (D) Trapping assays for GFP-DNMT1wt or GFP-DNMT1ΔE3–6 were performed as in C on three cells for each construct, and the estimated immobilized fractions were plotted with respect to the time of incubation with 5-aza-dC.
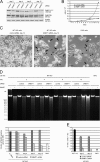
Figure 3.
Prolonged knockdown of DNMT1ΔE3–6 in MT1KO cells leads to genomic demethylation and cell death. Equal numbers of MT1KO cells were plated and transfected with either DNMT1 or control siRNA the next day (day 0) and every second day for 12 d. On days 2, 4, 8, and 12, cells were harvested, aliquots were collected for protein extracts and DNA isolation, and, except for day 12, equal numbers were replated. (A) Levels of DNMT1ΔE3–6 and DNMT3B were analyzed by Western blotting of whole cell extracts collected at the indicated days of siRNA treatment. Detection of β-actin was used to control for loading. (B) Cell numbers were plotted taking into consideration the splitting factor at each passage. (C) Phase-contrast images of MT1KO cultures treated with either control (left) or DNMT1 (middle) siRNA for 12 d and an image of DKO cells (right). Arrows point to dead cells, and arrowheads point to cells with thin and extended cytoplasmic protrusions that are present only in MT1KO cells treated with DNMT1 siRNA and in DKO cells. Bars, 50 μm. (D and E) Assays for the determination of global genomic methylation levels from the indicated cell lines and treatments. (D) Genomic DNA was digested with the McrBC endonuclease, which selectively recognizes methylated sequences. The top panel shows electrophoretic separation of the digests. Progressive demethylation in cells treated with DNMT1 siRNA is indicated by the increasingly similar patterns between mock digestions (−) and samples incubated with the enzyme (+). The bottom panel shows quantification of the McrBC-sensitive (methylated) DNA fraction from the samples shown in the top panel. (E) HPLC quantification of global 5-methyl-2′-deoxycytidine (mdC) content. Bars and error bars represent mean values and SEM from five measurements, respectively, except for HCT116 parental cells and MT1KO cells treated with DNMT1 siRNA for 12 d (six and two measurements, respectively).
Similar articles
- Dynamics of Dnmt1 interaction with the replication machinery and its role in postreplicative maintenance of DNA methylation.
Schermelleh L, Haemmer A, Spada F, Rösing N, Meilinger D, Rothbauer U, Cardoso MC, Leonhardt H. Schermelleh L, et al. Nucleic Acids Res. 2007;35(13):4301-12. doi: 10.1093/nar/gkm432. Epub 2007 Jun 18. Nucleic Acids Res. 2007. PMID: 17576694 Free PMC article. - The SRA protein Np95 mediates epigenetic inheritance by recruiting Dnmt1 to methylated DNA.
Sharif J, Muto M, Takebayashi S, Suetake I, Iwamatsu A, Endo TA, Shinga J, Mizutani-Koseki Y, Toyoda T, Okamura K, Tajima S, Mitsuya K, Okano M, Koseki H. Sharif J, et al. Nature. 2007 Dec 6;450(7171):908-12. doi: 10.1038/nature06397. Epub 2007 Nov 11. Nature. 2007. PMID: 17994007 - Suppression of clonogenicity by mammalian Dnmt1 mediated by the PCNA-binding domain.
Santourlidis S, Kimura F, Fischer J, Schulz WA. Santourlidis S, et al. Biochem Cell Biol. 2004 Oct;82(5):589-96. doi: 10.1139/o04-099. Biochem Cell Biol. 2004. PMID: 15499388 - Dnmt1 structure and function.
Svedružić ŽM. Svedružić ŽM. Prog Mol Biol Transl Sci. 2011;101:221-54. doi: 10.1016/B978-0-12-387685-0.00006-8. Prog Mol Biol Transl Sci. 2011. PMID: 21507353 Review. - Ras regulation of DNA-methylation and cancer.
Patra SK. Patra SK. Exp Cell Res. 2008 Apr 1;314(6):1193-201. doi: 10.1016/j.yexcr.2008.01.012. Epub 2008 Jan 26. Exp Cell Res. 2008. PMID: 18282569 Review.
Cited by
- Chromatin and DNA replication.
MacAlpine DM, Almouzni G. MacAlpine DM, et al. Cold Spring Harb Perspect Biol. 2013 Aug 1;5(8):a010207. doi: 10.1101/cshperspect.a010207. Cold Spring Harb Perspect Biol. 2013. PMID: 23751185 Free PMC article. Review. - Disruption of Dnmt1/PCNA/UHRF1 interactions promotes tumorigenesis from human and mice glial cells.
Hervouet E, Lalier L, Debien E, Cheray M, Geairon A, Rogniaux H, Loussouarn D, Martin SA, Vallette FM, Cartron PF. Hervouet E, et al. PLoS One. 2010 Jun 29;5(6):e11333. doi: 10.1371/journal.pone.0011333. PLoS One. 2010. PMID: 20613874 Free PMC article. - USP7 Is a Master Regulator of Genome Stability.
Valles GJ, Bezsonova I, Woodgate R, Ashton NW. Valles GJ, et al. Front Cell Dev Biol. 2020 Aug 5;8:717. doi: 10.3389/fcell.2020.00717. eCollection 2020. Front Cell Dev Biol. 2020. PMID: 32850836 Free PMC article. Review. - Antiproliferative effects of DNA methyltransferase 3B depletion are not associated with DNA demethylation.
Hagemann S, Kuck D, Stresemann C, Prinz F, Brueckner B, Mund C, Mumberg D, Sommer A. Hagemann S, et al. PLoS One. 2012;7(5):e36125. doi: 10.1371/journal.pone.0036125. Epub 2012 May 1. PLoS One. 2012. PMID: 22563479 Free PMC article. - Major and essential role for the DNA methylation mark in mouse embryogenesis and stable association of DNMT1 with newly replicated regions.
Takebayashi S, Tamura T, Matsuoka C, Okano M. Takebayashi S, et al. Mol Cell Biol. 2007 Dec;27(23):8243-58. doi: 10.1128/MCB.00899-07. Epub 2007 Sep 24. Mol Cell Biol. 2007. PMID: 17893328 Free PMC article.
References
- Baylin, S.B., and J.E. Ohm. 2006. Epigenetic gene silencing in cancer - a mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer. 6:107–116. - PubMed
- Bird, A. 2002. DNA methylation patterns and epigenetic memory. Genes Dev. 16:6–21. - PubMed
- Bonfils, C., N. Beaulieu, E. Chan, J. Cotton-Montpetit, and A.R. MacLeod. 2000. Characterization of the human DNA methyltransferase splice variant Dnmt1b. J. Biol. Chem. 275:10754–10760. - PubMed