YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA - PubMed (original) (raw)
YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA
Shuyun Dong et al. Mol Cell. 2007.
Abstract
Autoregulatory loops often provide precise control of the level of expression of specific genes that encode key regulatory proteins. Here we have defined a pathway by which Yra1p, a yeast mRNA export factor, controls its own expression. We show that YRA1 exon 1 sequences in cis and Yra1p in trans inhibit YRA1 pre-mRNA splicing and commit the pre-mRNA to nuclear export. Mex67p and Crm1p jointly promote YRA1 pre-mRNA export, and once in the cytoplasm, the pre-mRNA is degraded by a 5' to 3' decay mechanism that is dependent on the decapping activator Edc3p and on specific sequences in the YRA1 intron. These results illustrate how common steps in the nuclear processing, export, and degradation of a transcript can be uniquely combined to control the expression of a specific gene and suggest that Edc3p-mediated decay may have additional regulatory functions in eukaryotic cells.
Figures
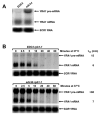
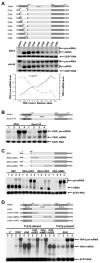
Figure 1. YRA1 Pre-mRNA is Selectively Stabilized in edc3Δ Cells
(A) Northern analysis of YRA1 pre-mRNA and mRNA levels in EDC3 and edc3Δ cells. (B) Determination of YRA1 pre-mRNA and mRNA half-lives. Cultures of wild-type and edc3Δ cells harboring the rpb1-1 allele were shifted from 25°C to 37°C to inactivate transcription by RNA polymerase II and samples were taken for northern analysis at different times after the shift. In panels A and B blots were hybridized with probes complementary to YRA1 and SCR1 transcripts, with the latter serving as a loading control.
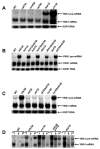
Figure 2. YRA1 Pre-mRNA is Degraded by Deadenylation-independent Decapping and 5′ to 3′ Exonucleolytic Digestion
(A–C) Northern analysis of YRA1 pre-mRNA and mRNA in yeast strains containing deletions of genes encoding: (A) the principal NMD factors, a subunit of the decapping enzyme, or the 5’ to 3’ exoribonuclease; (B) decapping activators of the general 5’ to 3’ mRNA decay pathway; or (C) components of the cytoplasmic exosome or the major cytoplasmic deadenylase complex. Blots were hybridized to a SCR1 probe to serve as a loading control. (D) Analysis of the 5’ cap status of the YRA1 pre-mRNA by anti-m7G immunoprecipitation and northern blotting. T: total input RNA, S: supernatant (uncapped), P: pellet (capped).
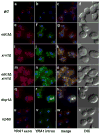
Figure 3. Edc3p-mediated YRA1 Pre-mRNA Degradation Occurs in the Cytoplasm
Localization of YRA1 pre-mRNA in wild-type, edc3Δ, xrn1Δ, xrn1Δedc3Δ, dcp1Δ, and rrp6Δ strains by in situ hybridization (FISH). Intron probes were labeled with Cy3 (green) and exon probes were labeled with Cy5 (red). DAPI labeling was used to identify the position of the nucleus. The corresponding merged and phase-contrast images are shown on the right side of the figure.
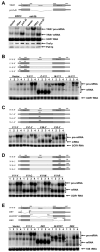
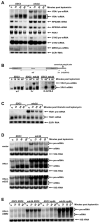
Figure 4. YRA1 Autoregulation Requires Edc3p and Two Distinct Regulatory Elements Localized in Exon1 and the Intron of YRA1 Pre-mRNA
(A) The effects of overexpressing intron-containing or intron-lacking YRA1 alleles on the levels of YRA1 pre-mRNA, mRNA, and Yra1p in EDC3 and edc3Δ strains. (B–D) Effects of replacing YRA1 exon1 (B), intron (C), and exon2 (D) with different sequences on YRA1 pre-mRNA and mRNA expression. A three-letter code was used to denote the order of exon1, intron, and exon2 of each chimeric construct, with the first letter of each of these genes specifying its origin. Y: YRA1, C: CYH2, M: MER2, R: RPS51A, and H: HIS3. (E) Effects of internal deletions in YRA1 exon1, intron, or exon2 on YRA1 pre-mRNA and mRNA expression. In panels B, C, D, and E YCp low-copy plasmids harboring the wild-type YRA1 gene, or a chimeric or deletion allele (depicted above the corresponding blots), were introduced into wild-type (1), upf1Δ (2), edc3Δ (3), or upf1Δ edc3Δ (4) strains and the levels of the respective pre-mRNAs and mRNAs encoded by these alleles were analyzed by northern blotting. The positions of chimeric or mutant pre-mRNAs and mRNAs are marked by Δ and *, respectively. Northern blots were hybridized to a SCR1 or an 18S rRNA probe to serve as loading controls. Pab1p served as a loading control for the western blot in panel A.
Figure 5. YRA1 Exon1 and Yra1p are cis- and _trans-_acting Negative Regulators of YRA1 Pre-mRNA Splicing
(A-D) Effects on the levels of YRA1 pre-mRNA and mRNA engendered by: (A) internal deletions in YRA1 exon1; (B) substituting YRA1 exon1 coding sequences with their complementary sequences; (C) mutations in the 5’ splice site, the 3’ splice site, or the branchpoint region of the YRA1 intron; and (D) mutations in the translation initiation codon, 5’ splice site, 3’ splice site, and branchpoint region of the YRA1 intron in the presence or absence of Yra1p. Note that, in panel B, the replaced complementary sequence does not contain in-frame premature stop codons. In panels A, B, C, and D plasmids carrying the wild-type YRA1 gene, or one of the mutant YRA1 alleles (depicted above the corresponding northern blots), were introduced into different yeast strains and the levels of the respective pre-mRNAs and mRNAs encoded by these alleles were analyzed by northern blotting. The relevant genotypes of yeast strains used in these experiments are indicated (panel A) or denoted by numbers in panels B, C, and D: 1: wild-type, 2: upf1Δ, 3_: edc3Δ,_ 4: upf1Δ edc3Δ, 5:EDC3 yra1Δ yra2Δ (YEplac112-YRA2), and 6: edc3Δ yra1Δ yra2Δ (YEplac112-YRA2). In panel A, the levels of mutant YRA1 pre-mRNAs and mRNAs in edc3Δ cells were quantified, normalized to the corresponding wild-type pre-mRNA and mRNA levels, and graphed below the northern blot. Blots were hybridized to a SCR1 probe to serve as a loading control.
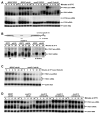
Figure 6. YRA1 Autoregulation Requires the Function of Mex67p
(A) Inactivation of Mex67p alters the levels of the YRA1, but not the CYH2 pre-mRNA and mRNA and also results in the accumulation of an atypical YRA1 RNA species in edc3Δ cells. (B) Inactivation of Mex67p promotes accumulation of YRA1 mRNA with an extended 3’-UTR. RNA samples from time points 0 and 24 min of the Mex67-5 EDC3 and Mex67-5 edc3Δ strains shown in panel A were analyzed with a YRA1 oligonucleotide probe that spans exon1 and exon2 (Left), an oligonucleotide probe downstream of the YRA1 translation termination codon (Middle), or an oligonucleotide probe downstream of the canonical poly(A) addition site (Right). (C) The effects of Mex67p inactivation on YRA1 mRNA level and the accumulation of YRA1 mRNA with an extended 3’-UTR requires ongoing transcription. (D) Deletion of UPF1, DCP1 or XRN1 restores YRA1 pre-mRNA levels at 37°C in mex67-5 edc3Δ cells. In panels A, C, and D, yeast strains of the indicated genotypes were grown at 25°C and then shifted to 37°C and the levels of YRA1 or CYH2 transcripts were analyzed by northern blotting. In panel C, thiolutin was added to cell cultures just before the temperature shift. A putative 3’ to 5’ decay intermediate of YRA1 pre-mRNA detected in the MEX67edc3Δ strain in panel A is marked by *. Blots were hybridized to a SCR1 probe to serve as a loading control.
Figure 7. Crm1p Mediates YRA1 Pre-mRNP Export to the Cytoplasm
(A) Inhibition of Crm1p function increases expression of YRA1 mRNA, but slightly decreases expression of other control RNAs including the CYH2 and MER3 pre-mRNAs, and the RPS28A, RPS28B, and PGK1 mRNAs (B) Inhibition of Crm1p promotes accumulation of YRA1 pre-mRNA and mRNA with extended 3’-UTRs. RNA samples from the indicated time points of the EDC3 and edc3Δ strains shown in panel A were analyzed with the same set of oligonucleotide probes used in Figure 6B. (C–D) Leptomycin-promoted increases in YRA1 mRNA levels and the accumulation of YRA1 mRNA with an extended 3’-UTR requires ongoing transcription (C) and YRA1 pre-mRNA splicing (D). (E) YRA1 pre-mRNA defective in splicing becomes a target of the nuclear exosome when Crm1p function is inhibited. In panels A, C, D, and E yeast strains of the indicated genotypes (all harboring the CRM1-T539C allele_)_ were treated with leptomycin and the effect on YRA1 pre-mRNA and mRNA were analyzed by northern blotting. In panel C, thiolutin and leptomycin were simultaneously added to EDC3 and edc3Δ cells. In panel D, a YCp plasmid harboring the YRA1 gene or its yra1-m5SS allele, as well as the empty vector, were introduced into EDC3 and edc3Δ strains. In panel E, a YCp plasmid harboring the yra1-mBB2 allele was introduced into yeast strains of the indicated genotypes. Blots were hybridized to a SCR1 or an 18S rRNA probe to serve as loading controls.
Similar articles
- Degradation of YRA1 Pre-mRNA in the cytoplasm requires translational repression, multiple modular intronic elements, Edc3p, and Mex67p.
Dong S, Jacobson A, He F. Dong S, et al. PLoS Biol. 2010 Apr 27;8(4):e1000360. doi: 10.1371/journal.pbio.1000360. PLoS Biol. 2010. PMID: 20463951 Free PMC article. - Expression of the essential mRNA export factor Yra1p is autoregulated by a splicing-dependent mechanism.
Preker PJ, Kim KS, Guthrie C. Preker PJ, et al. RNA. 2002 Aug;8(8):969-80. doi: 10.1017/s1355838202020046. RNA. 2002. PMID: 12212852 Free PMC article. - Perinuclear Mlp proteins downregulate gene expression in response to a defect in mRNA export.
Vinciguerra P, Iglesias N, Camblong J, Zenklusen D, Stutz F. Vinciguerra P, et al. EMBO J. 2005 Feb 23;24(4):813-23. doi: 10.1038/sj.emboj.7600527. Epub 2005 Feb 3. EMBO J. 2005. PMID: 15692572 Free PMC article. - [Coordinated pathway of nuclear mRNA export reveled by the genetic analysis in yeast].
Tani T. Tani T. Tanpakushitsu Kakusan Koso. 2009 Dec;54(16 Suppl):2102-8. Tanpakushitsu Kakusan Koso. 2009. PMID: 21089625 Review. Japanese. No abstract available. - Formation of export-competent mRNP: escaping nuclear destruction.
Saguez C, Olesen JR, Jensen TH. Saguez C, et al. Curr Opin Cell Biol. 2005 Jun;17(3):287-93. doi: 10.1016/j.ceb.2005.04.009. Curr Opin Cell Biol. 2005. PMID: 15901499 Review.
Cited by
- Yeast Edc3 targets RPS28B mRNA for decapping by binding to a 3' untranslated region decay-inducing regulatory element.
He F, Li C, Roy B, Jacobson A. He F, et al. Mol Cell Biol. 2014 Apr;34(8):1438-51. doi: 10.1128/MCB.01584-13. Epub 2014 Feb 3. Mol Cell Biol. 2014. PMID: 24492965 Free PMC article. - Crystal structure of human Edc3 and its functional implications.
Ling SH, Decker CJ, Walsh MA, She M, Parker R, Song H. Ling SH, et al. Mol Cell Biol. 2008 Oct;28(19):5965-76. doi: 10.1128/MCB.00761-08. Epub 2008 Aug 4. Mol Cell Biol. 2008. PMID: 18678652 Free PMC article. - Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development.
Xu J, Chua NH. Xu J, et al. Plant Cell. 2009 Oct;21(10):3270-9. doi: 10.1105/tpc.109.070078. Epub 2009 Oct 23. Plant Cell. 2009. PMID: 19855049 Free PMC article. - Decapping activators Edc3 and Scd6 act redundantly with Dhh1 in post-transcriptional repression of starvation-induced pathways.
Kumar R, Zhang F, Niphadkar S, Onu C, Vijjamarri AK, Greenberg ML, Laxman S, Hinnebusch AG. Kumar R, et al. bioRxiv [Preprint]. 2024 Aug 28:2024.08.28.610059. doi: 10.1101/2024.08.28.610059. bioRxiv. 2024. PMID: 39257769 Free PMC article. Preprint. - RNA degradation in Saccharomyces cerevisae.
Parker R. Parker R. Genetics. 2012 Jul;191(3):671-702. doi: 10.1534/genetics.111.137265. Genetics. 2012. PMID: 22785621 Free PMC article. Review.
References
- Badis G, Saveanu C, Fromont-Racine M, Jacquier A. Targeted mRNA Degradation by Deadenylation-Independent Decapping. Mol Cell. 2004;15:5–15. - PubMed
- Bousquet-Antonelli C, Presutti C, Tollervey D. Identification of a regulated pathway for nuclear pre-mRNA turnover. Cell. 2000;102:765–775. - PubMed
- Coller J, Parker R. Eukaryotic mRNA decapping. Annu Rev Biochem. 2004;73:861–890. - PubMed
- Cullen BR. Nuclear mRNA export: insights from virology. Trends Biochem Sci. 2003;28:419–424. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 GM057071/GM/NIGMS NIH HHS/United States
- GM57071/GM/NIGMS NIH HHS/United States
- R01 GM027757/GM/NIGMS NIH HHS/United States
- R37 GM027757/GM/NIGMS NIH HHS/United States
- GM27757/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases