Genomic relationships and speciation times of human, chimpanzee, and gorilla inferred from a coalescent hidden Markov model - PubMed (original) (raw)
Genomic relationships and speciation times of human, chimpanzee, and gorilla inferred from a coalescent hidden Markov model
Asger Hobolth et al. PLoS Genet. 2007.
Abstract
The genealogical relationship of human, chimpanzee, and gorilla varies along the genome. We develop a hidden Markov model (HMM) that incorporates this variation and relate the model parameters to population genetics quantities such as speciation times and ancestral population sizes. Our HMM is an analytically tractable approximation to the coalescent process with recombination, and in simulations we see no apparent bias in the HMM estimates. We apply the HMM to four autosomal contiguous human-chimp-gorilla-orangutan alignments comprising a total of 1.9 million base pairs. We find a very recent speciation time of human-chimp (4.1 +/- 0.4 million years), and fairly large ancestral effective population sizes (65,000 +/- 30,000 for the human-chimp ancestor and 45,000 +/- 10,000 for the human-chimp-gorilla ancestor). Furthermore, around 50% of the human genome coalesces with chimpanzee after speciation with gorilla. We also consider 250,000 base pairs of X-chromosome alignments and find an effective population size much smaller than 75% of the autosomal effective population sizes. Finally, we find that the rate of transitions between different genealogies correlates well with the region-wide present-day human recombination rate, but does not correlate with the fine-scale recombination rates and recombination hot spots, suggesting that the latter are evolutionarily transient.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures
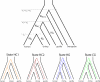
Figure 1. Genetic and Species Relationships May Differ
Top: Genealogical relationship of human, chimpanzee, gorilla, and orangutan. Speciation times are denoted τ 1, τ 1 + τ 2, and τ 1 + τ 2 + τ 3. Population sizes of human, chimpanzee, and gorilla are denoted N H, N C, and N G, while the HC and HCG ancestral population sizes are denoted N HC and N HCG. Bottom: Each of the four hidden states in the coal-HMM corresponds to a particular phylogenetic tree. In state HC1, human and chimpanzee coalesce before speciation of human, chimpanzee, and gorilla, i.e., before τ 1 + τ 2. In states HC2, HG, and CG, human, chimpanzee, and gorilla coalesce after speciation of the three species, i.e., after τ 1 + τ 2. In HC2, the human and chimpanzee lineages coalesce first, and then the HC lineage coalesces with gorilla. In state HG, human and gorilla coalesce first, and in state CG, chimpanzee and gorilla coalesce first. The hidden phylogenetic states cannot be observed from present-day sequence data, but they can be decoded using the coal-HMM methodology.
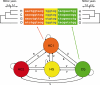
Figure 2. Graphical Representation of Coal-HMM
The coal-HMM divides a multiple alignment into four types of segments corresponding to the four phylogenetic states HC1, HC2, HG, and CG. The probability of making a transition from state HC1 to any of the other states is s, and the probability of a transition from any of the HC2, HG, or CG states to state HC1 is u. Transitions between the HC2, HG, and CG states have probability v. The indicated branch lengths of the phylogenetic trees from state HC1 and state CG are divergence times estimated from Target 1. The branch lengths of the phylogenetic trees corresponding to state HC2 and HG are the same as the branch lengths of state CG.
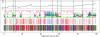
Figure 3. Inferred Genealogies from Real Data
(From bottom to top) Analysis of the first 100 kb from Target 1. (Bottom) Site information without outgroup: sites shared by HC in red, by HG in blue, and by CG in green (compare first, third, and last columns in Table 1). (Second from bottom) Site information with outgroup: sites strongly supporting states HC1 or HC2 in red, HG in blue, and CG in green (compare second, third, and last columns in Table 1). (Third from bottom) Posterior probabilities: Coloring as in second from bottom, except that state HC2 is dark red. (Top) Fine-scale recombination rate estimates (log scale). The vertical lines mark subdivisions of the multiple alignment due to more than 50-base-pair deletions in one species (see “Data” in Materials and Methods).
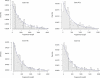
Figure 4. Parameter Estimates from Coal-HMM Analysis of Five Targets
Estimates with associated standard errors of the HMM and population genetics parameters for the five targets. Top left plot shows the HMM transition rates, top right plot the genetic divergence times in million years (assuming orangutan divergence 18 Myr ago), bottom left plot the speciation times in million years, and bottom right plot the ancestral effective population sizes, again assuming orangutan divergence of 18 Myr and a generation time of 25 y for all species throughout the HCGO divergence.
Figure 5. Inferred Genealogies along a 6-kb Region of the X Chromosome
We observe several adjacent sites that support alternative state HG (blue lines), corresponding to coalescence of human and gorilla before coalescence with chimpanzee.
Figure 6. Relation between Parameters in the Coal-HMM and Coalescent-with-Recombination Parameters
Left: Coal-HMM and coalescent parameters in state HC1. Right: Coal-HMM and coalescent parameters in state HC2. In both states we assume a molecular clock.
Figure 7. Histogram of Fragment Lengths for the Four Different Genealogies
The distribution of fragment lengths is reasonably well approximated by the geometric distribution (blue line). This property is a basic assumption of the coal-HMM.
Figure 8. Inferred Genealogies from Simulated Data
(Bottom) Site information without outgroup: sites shared by HC in red, by HG in blue, and by CG in green (see first, third, and last columns in Table 1). (Second from bottom) True genealogical state in simulations: state HC1 in red, HC2 in dark red, HG in blue, and CG in green. (Third from bottom) Site information with outgroup: sites supporting state HC1 or HC2 in red, HG in blue, and CG in green (see second, third, and last columns in Table 1). (Top) Posterior probabilities for the four states. Coloring as in second from bottom.
Similar articles
- Ancestral population genomics: the coalescent hidden Markov model approach.
Dutheil JY, Ganapathy G, Hobolth A, Mailund T, Uyenoyama MK, Schierup MH. Dutheil JY, et al. Genetics. 2009 Sep;183(1):259-74. doi: 10.1534/genetics.109.103010. Epub 2009 Jul 6. Genetics. 2009. PMID: 19581452 Free PMC article. - Reconstructing the demographic history of the human lineage using whole-genome sequences from human and three great apes.
Hara Y, Imanishi T, Satta Y. Hara Y, et al. Genome Biol Evol. 2012;4(11):1133-45. doi: 10.1093/gbe/evs075. Genome Biol Evol. 2012. PMID: 22975719 Free PMC article. - A new isolation with migration model along complete genomes infers very different divergence processes among closely related great ape species.
Mailund T, Halager AE, Westergaard M, Dutheil JY, Munch K, Andersen LN, Lunter G, Prüfer K, Scally A, Hobolth A, Schierup MH. Mailund T, et al. PLoS Genet. 2012;8(12):e1003125. doi: 10.1371/journal.pgen.1003125. Epub 2012 Dec 20. PLoS Genet. 2012. PMID: 23284294 Free PMC article. - TRAILS: Tree reconstruction of ancestry using incomplete lineage sorting.
Rivas-González I, Schierup MH, Wakeley J, Hobolth A. Rivas-González I, et al. PLoS Genet. 2024 Feb 8;20(2):e1010836. doi: 10.1371/journal.pgen.1010836. eCollection 2024 Feb. PLoS Genet. 2024. PMID: 38330138 Free PMC article. - [Molecular phylogeny of humans, chimpanzees and gorillas].
Satta Y. Satta Y. Tanpakushitsu Kakusan Koso. 2000 Dec;45(16):2588-95. Tanpakushitsu Kakusan Koso. 2000. PMID: 11185912 Review. Japanese. No abstract available.
Cited by
- Population genetic studies in the genomic sequencing era.
Chen H. Chen H. Dongwuxue Yanjiu. 2015 Jul 18;36(4):223-32. doi: 10.13918/j.issn.2095-8137.2015.4.223. Dongwuxue Yanjiu. 2015. PMID: 26228473 Free PMC article. Review. - Molecular phylogenetic analysis of non-sexually transmitted strains of Haemophilus ducreyi.
Gaston JR, Roberts SA, Humphreys TL. Gaston JR, et al. PLoS One. 2015 Mar 16;10(3):e0118613. doi: 10.1371/journal.pone.0118613. eCollection 2015. PLoS One. 2015. PMID: 25774793 Free PMC article. - Molecular phylogenetics: principles and practice.
Yang Z, Rannala B. Yang Z, et al. Nat Rev Genet. 2012 Mar 28;13(5):303-14. doi: 10.1038/nrg3186. Nat Rev Genet. 2012. PMID: 22456349 Review. - Proboscidean mitogenomics: chronology and mode of elephant evolution using mastodon as outgroup.
Rohland N, Malaspinas AS, Pollack JL, Slatkin M, Matheus P, Hofreiter M. Rohland N, et al. PLoS Biol. 2007 Aug;5(8):e207. doi: 10.1371/journal.pbio.0050207. PLoS Biol. 2007. PMID: 17676977 Free PMC article. - Natural selection affects multiple aspects of genetic variation at putatively neutral sites across the human genome.
Lohmueller KE, Albrechtsen A, Li Y, Kim SY, Korneliussen T, Vinckenbosch N, Tian G, Huerta-Sanchez E, Feder AF, Grarup N, Jørgensen T, Jiang T, Witte DR, Sandbæk A, Hellmann I, Lauritzen T, Hansen T, Pedersen O, Wang J, Nielsen R. Lohmueller KE, et al. PLoS Genet. 2011 Oct;7(10):e1002326. doi: 10.1371/journal.pgen.1002326. Epub 2011 Oct 13. PLoS Genet. 2011. PMID: 22022285 Free PMC article.
References
- Enard W, Pääbo S. Comparative primate genomics. Annu Rev Genomics Hum Genet. 2004;5:351–378. - PubMed
- Patterson N, Richter DJ, Gnerre S, Lander ES, Reich D. Genetic evidence for complex speciation of human and chimpanzees. Nature. 2006;441:1103–1108. - PubMed
- Innan H, Watanabe H. The effect of gene flow on the coalescent time in the human-chimpanzee ancestral population. Mol Biol Evol. 2006;23:1040–1047. - PubMed
- Barton NH. Evolutionary biology: How did the human species form? Curr Biol. 2006;16:R647–R650. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources