Design, expression, and stability of a diverse protein library based on the human fibronectin type III domain - PubMed (original) (raw)
Design, expression, and stability of a diverse protein library based on the human fibronectin type III domain
C Anders Olson et al. Protein Sci. 2007 Mar.
Abstract
Protein libraries based on natural scaffolds enable the generation of novel molecular tools and potential therapeutics by directed evolution. Here, we report the design and construction of a high complexity library (30 x 10(13) sequences) based on the 10th fibronectin type III domain of human fibronectin (10FnIII). We examined the bacterial expression characteristics and stability of this library using a green fluorescent protein (GFP)-reporter screen, SDS-PAGE analysis, and chemical denaturation, respectively. The high throughput GFP reporter screen demonstrates that a large fraction of our library expresses significant levels of soluble protein in bacteria. However, SDS-PAGE analysis of expression cultures indicates the ratio of soluble to insoluble protein expressed varies greatly for randomly chosen library members. We also tested the stabilities of several representative variants by guanidinium chloride denaturation. All variants tested displayed cooperative unfolding transitions similar to wild-type, and two exhibited free energies of unfolding equal to wild-type 10FnIII. This work demonstrates the utility of GFP-based screening as a tool for analysis of high-complexity protein libraries. Our results indicate that a vast amount of protein sequence space surrounding the 10FnIII scaffold is accessible for the generation of novel functions by directed as well as natural evolution.
Figures
Figure 1.
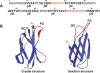
Sequence and structure of 10FnIII. (A) Sequence of 10FnIII. Residues that are part of the β-strand framework are in bold. Residue positions that were selected for randomization are colored red. (B) Two structural views of 10FnIII. The crystal structure of the 10FnIII domain shown is part of the 7–10FnIII domains (left, PDB accession no. 1FNF). The randomized BC and FG loop regions are colored red. The first seven N-terminal residues are colored green. The solution structure of 10FnIII is illustrated in a side view without the unstructured N-terminal residues (right, PDB accession no. 1TTG).
Figure 2.
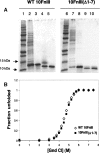
Purification and stability of WT 10FnIII compared to 10FnIII(Δ1–7). (A) Purification of WT 10FnIII (lanes 1–5) and 10FnIII(Δ1–7) (lanes 6–10). (Lanes 1,6) Protein standard. (Lanes 2,7) Cleared lysate. (Lanes 3,8) HisTrap column purification. (Lanes 4,9) Factor Xa cleavage of His6-tag. (Lanes 5,10) Gel filtration purification. (B) Guanidinium chloride denaturation of WT 10FnIII (open diamonds) and 10FnIII(Δ1–7) (closed squares) monitored by Trp fluorescence.
Figure 3.
Scheme for constructing 10FnIII library. Linear illustration of library DNA (A) generated from eight oligonucleotides (B).
Figure 4.
Random sequence composition. The frequencies of each amino acid (closed diamonds) are compared to the target NNS distribution (open circles) and the composition of natural protein interfaces involved in protein–protein binding (Chakrabarti and Janin 2002).
Figure 5.
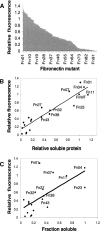
Expression analysis of 10FnIII library. (A) Expression fitness landscape of the 10FnIII library. Fluorescence intensity values of 94 Fn variant–GFP fusions were obtained from cell suspensions and normalized to WT 10FnIII(Δ1–7)-GFP. Each variant is marked by a dash on the X axis. (B) Relation between relative fluorescence and Fn variant soluble expression. Nineteen variants plus WT 10FnIII(Δ1–7) were subcloned into an expression vector without GFP. The relative expression values represent the amount of protein expressed in the soluble fraction relative to wild type measured by band densitometry. (C) The relation between relative fluorescence and Fn variant solubility, determined by band densitometry of soluble and insoluble fractions.
Figure 6.
Guanidinium chloride denaturation of 10FnIII variants compared to WT10 FnIII(Δ1–7). (Closed triangles) Fn04; (open circles) Fn23; (open squares) Fn32; (closed circles) Fn38; (crosses) WT 10FnIII(Δ1–7).
Similar articles
- Mechanical unfolding intermediates observed by single-molecule force spectroscopy in a fibronectin type III module.
Li L, Huang HH, Badilla CL, Fernandez JM. Li L, et al. J Mol Biol. 2005 Jan 28;345(4):817-26. doi: 10.1016/j.jmb.2004.11.021. J Mol Biol. 2005. PMID: 15588828 - Interdomain mobility and conformational stability of type III fibronectin domain pairs control surface adsorption, desorption and unfolding.
Pereira P, Kelly SM, Gellert PR, van der Walle CF. Pereira P, et al. Colloids Surf B Biointerfaces. 2008 Jun 15;64(1):1-9. doi: 10.1016/j.colsurfb.2007.12.015. Epub 2008 Jan 3. Colloids Surf B Biointerfaces. 2008. PMID: 18261887 - Monobodies: antibody mimics based on the scaffold of the fibronectin type III domain.
Koide A, Koide S. Koide A, et al. Methods Mol Biol. 2007;352:95-109. doi: 10.1385/1-59745-187-8:95. Methods Mol Biol. 2007. PMID: 17041261 - Evolution of an arbitrary sequence in solubility.
Ito Y, Kawama T, Urabe I, Yomo T. Ito Y, et al. J Mol Evol. 2004 Feb;58(2):196-202. doi: 10.1007/s00239-003-2542-2. J Mol Evol. 2004. PMID: 15042340 - Combinatorial library approaches for improving soluble protein expression in Escherichia coli.
Hart DJ, Tarendeau F. Hart DJ, et al. Acta Crystallogr D Biol Crystallogr. 2006 Jan;62(Pt 1):19-26. doi: 10.1107/S0907444905036097. Epub 2005 Dec 14. Acta Crystallogr D Biol Crystallogr. 2006. PMID: 16369090 Review.
Cited by
- Creating stable stem regions for loop elongation in Fcabs - insights from combining yeast surface display, in silico loop reconstruction and molecular dynamics simulations.
Hasenhindl C, Lai B, Delgado J, Traxlmayr MW, Stadlmayr G, Rüker F, Serrano L, Oostenbrink C, Obinger C. Hasenhindl C, et al. Biochim Biophys Acta. 2014 Sep;1844(9):1530-40. doi: 10.1016/j.bbapap.2014.04.020. Epub 2014 May 2. Biochim Biophys Acta. 2014. PMID: 24792385 Free PMC article. - The HCM-linked W792R mutation in cardiac myosin-binding protein C reduces C6 FnIII domain stability.
Smelter DF, de Lange WJ, Cai W, Ge Y, Ralphe JC. Smelter DF, et al. Am J Physiol Heart Circ Physiol. 2018 Jun 1;314(6):H1179-H1191. doi: 10.1152/ajpheart.00686.2017. Epub 2018 Feb 16. Am J Physiol Heart Circ Physiol. 2018. PMID: 29451820 Free PMC article. - An E3-ligase-based method for ablating inhibitory synapses.
Gross GG, Straub C, Perez-Sanchez J, Dempsey WP, Junge JA, Roberts RW, Trinh le A, Fraser SE, De Koninck Y, De Koninck P, Sabatini BL, Arnold DB. Gross GG, et al. Nat Methods. 2016 Aug;13(8):673-8. doi: 10.1038/nmeth.3894. Epub 2016 Jun 6. Nat Methods. 2016. PMID: 27271196 Free PMC article. - Mammalian Expression and In Situ Biotinylation of Extracellular Protein Targets for Directed Evolution.
Grindel BJ, Engel BJ, Hall CG, Kelderhouse LE, Lucci A, Zacharias NM, Takahashi TT, Millward SW. Grindel BJ, et al. ACS Omega. 2020 Sep 22;5(39):25440-25455. doi: 10.1021/acsomega.0c03990. eCollection 2020 Oct 6. ACS Omega. 2020. PMID: 33043224 Free PMC article. - Extremophilic 50S Ribosomal RNA-Binding Protein L35Ae as a Basis for Engineering of an Alternative Protein Scaffold.
Lomonosova AV, Ovchinnikova EV, Kazakov AS, Denesyuk AI, Sofin AD, Mikhailov RV, Ulitin AB, Mirzabekov TA, Permyakov EA, Permyakov SE. Lomonosova AV, et al. PLoS One. 2015 Aug 6;10(8):e0134906. doi: 10.1371/journal.pone.0134906. eCollection 2015. PLoS One. 2015. PMID: 26247602 Free PMC article.
References
- Batori V., Koide, A., and Koide, S. 2002. Exploring the potential of the monobody scaffold: Effects of loop elongation on the stability of a fibronectin type III domain. Protein Eng. 15: 1015–1020. - PubMed
- Binz H.K. and Pluckthun, A. 2005. Engineered proteins as specific binding reagents. Curr. Opin. Biotechnol. 16: 459–469. - PubMed
- Binz H.K., Stumpp, M.T., Forrer, P., Amstutz, P., and Pluckthun, A. 2003. Designing repeat proteins: Well-expressed, soluble and stable proteins from combinatorial libraries of consensus ankyrin repeat proteins. J. Mol. Biol. 332: 489–503. - PubMed
- Binz H.K., Amstutz, P., Kohl, A., Stumpp, M.T., Briand, C., Forrer, P., Grutter, M.G., and Pluckthun, A. 2004. High-affinity binders selected from designed ankyrin repeat protein libraries. Nat. Biotechnol. 22: 575–582. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources